Harnessing single-cell and multi-omics insights: STING pathway-based predictive signature for immunotherapy response in lung adenocarcinoma
- PMID: 40308576
- PMCID: PMC12040650
- DOI: 10.3389/fimmu.2025.1575084
Harnessing single-cell and multi-omics insights: STING pathway-based predictive signature for immunotherapy response in lung adenocarcinoma
Erratum in
-
Corrigendum: Harnessing single-cell and multi-omics insights: STING pathway-based predictive signature for immunotherapy response in lung adenocarcinoma.Front Immunol. 2025 Apr 30;16:1613095. doi: 10.3389/fimmu.2025.1613095. eCollection 2025. Front Immunol. 2025. PMID: 40370448 Free PMC article.
Abstract
Background: Lung adenocarcinoma is the most prevalent type of small-cell carcinoma, with a poor prognosis. For advanced-stage patients, the efficacy of immunotherapy is suboptimal. The STING signaling pathway plays a pivotal role in the immunotherapy of lung adenocarcinoma; therefore, further investigation into the relationship between the STING pathway and lung adenocarcinoma is warranted.
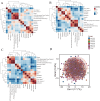
Methods: We conducted a comprehensive analysis integrating single-cell RNA sequencing (scRNA-seq) data with bulk transcriptomic profiles from public databases (GEO, TCGA). STING pathway-related genes were identified through Genecard database. Advanced bioinformatics analyses using R packages (Seurat, CellChat) revealed transcriptomic heterogeneity, intercellular communication networks, and immune landscape characteristics. We developed a STING pathway-related signature (STINGsig) using 101 machine learning frameworks. The functional significance of ERRFI1, a key component of STINGsig, was validated through mouse models and multicolor flow cytometry, particularly examining its role in enhancing antitumor immunity and potential synergy with α-PD1 therapy.
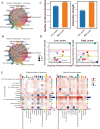
Results: Our single-cell analysis identified and characterized 15 distinct cell populations, including epithelial cells, macrophages, fibroblasts, T cells, B cells, and endothelial cells, each with unique marker gene profiles. STING pathway activity scoring revealed elevated activation in neutrophils, epithelial cells, B cells, and T cells, contrasting with lower activity in inflammatory macrophages. Cell-cell communication analysis demonstrated enhanced interaction networks in high-STING-score cells, particularly evident in fibroblasts and endothelial cells. The developed STINGsig showed robust prognostic value and revealed distinct immune microenvironment characteristics between risk groups. Notably, ERRFI1 knockdown experiments confirmed its significant role in modulating antitumor immunity and enhancing α-PD1 therapy response.
Conclusion: The STING-related pathway exhibited distinct expression levels across 15 cell populations, with high-score cells showing enhanced tumor-promoting pathways, active immune interactions, and enrichment in fibroblasts and IFI27+ inflammatory macrophages. In contrast, low-score cells were associated with epithelial phenotypes and reduced immune activity. We developed a robust STING pathway-related signature (STINGsig), which identified key prognostic genes and was linked to the immune microenvironment. Through in vivo experiments, we confirmed that knockdown of ERRFI1, a critical gene within the STINGsig, significantly enhances antitumor immunity and synergizes with α-PD1 therapy in a lung cancer model, underscoring its therapeutic potential in modulating immune responses.
Keywords: lung adenocarcinoma; machine learning; non-small cell lung cancer; prognosis signature; single-cell RNA sequencing.
Copyright © 2025 Ding, Wang, Yan, Fan, Ding and Xue.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

