Regulatory element map of sheep reproductive tissues: functional annotation of tissue-specific strong active enhancers
- PMID: 40308692
- PMCID: PMC12040938
- DOI: 10.3389/fvets.2025.1564148
Regulatory element map of sheep reproductive tissues: functional annotation of tissue-specific strong active enhancers
Abstract
Introduction: Comprehensive functional annotation of the genome is crucial for elucidating the molecular mechanisms underlying complex traits and diseases. Although functional annotation has been partially completed in sheep, a systematic annotation focused on reproductive tissues remains absent.
Methods: In this study, we integrated 60 transcriptomic and epigenomic datasets from five reproductive tissues. Using a multi-omics approach, we predicted 15 distinct chromatin states and conducted thorough functional annotation.
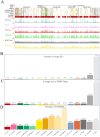
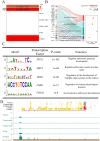
Results: We established the first regulatory element atlas for sheep reproductive tissues and examined the roles of these elements in reproductive traits and disease. In total, we annotated 1,680,172 regulatory elements, including 83,980 tissue-specific strong active enhancers (EnhAs).
Discussion: Enhancers were identified as critical drivers of tissue-specific functions, operating through sequence-specific transcription factor binding and direct regulation of target genes. Key transcription factors associated with reproductive function included INHBA (ovary), KITLG (oviduct), Snai2 (cervix), WNT7A (uterine horn), FOLR1 (uterine body), and SALL1 (shared uterine regions). Additionally, our findings support the potential of sheep as a promising model for investigating embryonic development and miscarriage. This work lays a theoretical foundation for future research into the molecular mechanisms of complex traits and diseases in sheep.
Keywords: enhancer; regulatory element; reproductive tissues; sheep; tissue specific.
Copyright © 2025 Meng, Chu, Yang, Zhang, Wang, Chen, Ren, Pan and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Mazinani M, Rude B. Population, world production and quality of sheep and goat products. Am J Anim Vet Sci. (2020) 15:291–9. doi: 10.3844/ajavsp.2020.291.299 - DOI
-
- Gonçalves JD, Dias JH, Machado-Neves M, Vergani GB, Ahmadi B, Pereira Batista RIT, et al. . Transcervical uterine flushing and embryo transfer in sheep: Morphophysiological basis for approaches currently used, major challenges, potential improvements, and new directions (alas, including some old ideas). Reprod Biol. (2024) 24:100920. doi: 10.1016/j.repbio.2024.100920, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

