Inborn errors of immunity underlie clonal T cell expansions in large granular lymphocyte leukemia
- PMID: 40309770
- PMCID: PMC12043085
- DOI: 10.1172/JCI184431
Inborn errors of immunity underlie clonal T cell expansions in large granular lymphocyte leukemia
Abstract
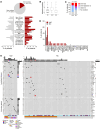
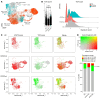
BACKGROUNDT cell large granular lymphocyte leukemia (T-LGLL) is a lymphoproliferative disorder of cytotoxic T lymphocytes (CTLs), often with gain-of-function STAT3 mutations. T-LGLL represents a unique model for the study of persistent CTL expansions. Albeit autoimmunity is implied, various paradoxical observations led us to investigate whether immunodeficiency traits underpin T-LGLL.METHODSThis is a comprehensive immunogenomic study of 92 consecutive patients from a large T-LGLL cohort with full laboratory-clinical characterization (n = 271). Whole-exome profiling of variants associated with inborn errors of immunity (IEI) and somatic mutations in T cell lymphoid drivers was analyzed. Single-cell RNA-Seq and TCR-Seq in T-LGLL samples and RNA-Seq in T cell cancer cell lines were utilized to establish biological correlations.RESULTSLymphocytopenia and/or hypogammaglobulinemia were identified in 186 of 241 (77%) T-LGLL patients. Genetic screening for IEI revealed 43 rare heterozygous variants in 38 different immune genes in 34 of 92 (36%) patients (vs. 167/63,026 [0.26%] in controls). High-confidence deleterious variants associated with dominant, adult-onset IEIs were detected in 15 of 92 (16%) patients. Carriers showed atypical features otherwise tied to the cryptic IEI, such as earlier onset, lower lymphocyte counts, lower STAT3 mutational rate, and higher proportions of hypogammaglobulinemia and immune cytopenia/bone marrow failure than noncarriers. Somatic mutational landscape, RNA-Seq, and TCR-Seq analyses supported immune imbalance caused by the IEI variants and interactions with somatic mutations in T cell lymphoid drivers.CONCLUSIONSOur findings in T-LGLL reveal that maladaptive CTL expansions may stem from cryptic immunodeficiency traits and open the horizon of IEIs to clonal hematopoiesis and bone marrow failure.FUNDINGNIH; Aplastic Anemia and MDS International Foundation; VeloSano; Edward P. Evans Foundation; Instituto de Salud Carlos III; European Research Council; European Research Area Network on Personalised Medicine; Academy Finland; Cancer Foundation Finland.
Keywords: Adaptive immunity; Genetics; Hematology; Lymphomas; Molecular genetics.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

