Genome-wide association analysis identifies APOE as a mitophagy modifier in Lewy body disease
- PMID: 40309932
- PMCID: PMC12044520
- DOI: 10.1002/alz.70198
Genome-wide association analysis identifies APOE as a mitophagy modifier in Lewy body disease
Abstract
Introduction: Phosphorylated ubiquitin (p-S65-Ub) is generated during PINK1-PRKN mitophagy as a specific marker of mitochondrial damage. Despite the widespread deposition of p-S65-Ub in aged and diseased human brain, the genetic contribution to its accumulation remains unclear.
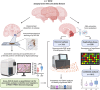
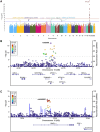
Methods: To identify novel mitophagy regulators, we performed a genome-wide association study using p-S65-Ub level as a quantitative trait in 1012 autopsy-confirmed Lewy body disease (LBD) samples.
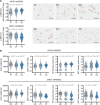
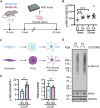
Results: We identified a significant genome-wide association with p-S65-Ub for rs429358 (apolipoprotein E ε4 [APOE4]) and a suggestive association for rs6480922 (ZMIZ1). APOE4 was associated with higher p-S65-Ub levels and greater neuropathological burden. Functional validation in mouse and human induced pluripotent stem cell (iPSC) models confirmed APOE4-mediated mitophagy alterations. Intriguingly, ZMIZ1 rs6480922 was associated with lower p-S65-Ub levels, reduced neuropathological load, and increased brain weight, indicating a potential protective role.
Discussion: Our findings underscore the importance of mitochondrial quality control in LBD pathogenesis and nominate regulators that may contribute to disease risk or resilience.
Highlights: p-S65-Ub levels were used as a quantitative marker of mitochondrial damage. A GWAS identified two genetic variants that modify mitophagy in LBD autopsy brain. APOE4 was associated with increased p-S65-Ub accumulation and neuropathology. APOE4 altered mitophagy via pathology-dependent and pathology-independent mechanisms. ZMIZ1 was linked to reduced p-S65-Ub and neuropathology indicative of protection.
Keywords: GWAS; PINK1; PRKN; Parkin; Parkinson's disease; ZMIZ1; autophagy; mitochondria; phosphorylated ubiquitin; ubiquitin.
© 2025 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
Mayo Clinic, FCF, and WS hold a patent related to PRKN activators (Small Molecule Activators of Parkin Enzyme Function, US Patent 11401255B2; August 2, 2022). All other authors declare they have no competing interests. This research was conducted in compliance with Mayo Clinic conflict of interest policies. Author disclosures are available in the Supporting Information.
Figures
Update of
-
Genome-wide association study identifies APOE and ZMIZ1 variants as mitophagy modifiers in Lewy body disease.medRxiv [Preprint]. 2023 Oct 16:2023.10.16.23297100. doi: 10.1101/2023.10.16.23297100. medRxiv. 2023. Update in: Alzheimers Dement. 2025 Apr;21(4):e70198. doi: 10.1002/alz.70198. PMID: 37905059 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- Ted Turner and Family
- U54NS110435/NS/NINDS NIH HHS/United States
- RF1AG046205/AG/NIA NIH HHS/United States
- AARF-22-973152/ALZ/Alzheimer's Association/United States
- P50 NS072187/NS/NINDS NIH HHS/United States
- The Ted Nash Long Life Foundation
- W81XWH-17-1-0248/Congressionally Directed Medical Research Programs
- U01NS100620/NS/NINDS NIH HHS/United States
- R01 NS078086/NS/NINDS NIH HHS/United States
- R01 AG056366/AG/NIA NIH HHS/United States
- The Little Family Foundation
- The Michael J. Fox Foundation for Parkinson's Research
- R01NS078086/NS/NINDS NIH HHS/United States
- R01 AG089380/AG/NIA NIH HHS/United States
- U19AG069701/AG/NIA NIH HHS/United States
- P30AG062677/AG/NIA NIH HHS/United States
- U54NS100693/NS/NINDS NIH HHS/United States
- P50NS072187/NS/NINDS NIH HHS/United States
- American Brain Foundation
- Mayo Clinic Center for Biomedical Discovery
- R01NS110085/NS/NINDS NIH HHS/United States
- Mayo Clinic Office of Research Equity, Inclusion, and Diversity
- Mayo Clinic Robert and Arlene Kogod Center on Aging
- 22A05/Florida Department of Health-Ed, Ethel Moore Alzheimer's Disease Research Program
- R01AG056366/AG/NIA NIH HHS/United States
- U54 NS110435/NS/NINDS NIH HHS/United States
- RF1 NS085070/NS/NINDS NIH HHS/United States
- U19 AG071754/AG/NIA NIH HHS/United States
- W81XWH-17-1-0249/Congressionally Directed Medical Research Programs
- U19AG071754/AG/NIA NIH HHS/United States
- P30 AG062677/AG/NIA NIH HHS/United States
- RF1 AG046205/AG/NIA NIH HHS/United States
- Mangurian Foundation Lewy Body Dementia Program at Mayo Clinic
- U19 AG069701/AG/NIA NIH HHS/United States
- R56AG062556/AG/NIA NIH HHS/United States
- RF1NS085070/NS/NINDS NIH HHS/United States
- R01AG089380/AG/NIA NIH HHS/United States
- R56 AG062556/AG/NIA NIH HHS/United States
- 22A07/Florida Department of Health-Ed, Ethel Moore Alzheimer's Disease Research Program
- U01 NS100620/NS/NINDS NIH HHS/United States
- R01 NS110085/NS/NINDS NIH HHS/United States
- American Parkinson Disease Association Center for Advanced Research at Mayo Clinic Jacksonville
- R01AG087165/AG/NIA NIH HHS/United States
- U54 NS100693/NS/NINDS NIH HHS/United States
- R01 AG087165/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

