Genomics-based approach for detection and characterization of SARS-CoV-2 co-infections and diverse viral populations
- PMID: 40310264
- PMCID: PMC12131723
- DOI: 10.1128/spectrum.02092-24
Genomics-based approach for detection and characterization of SARS-CoV-2 co-infections and diverse viral populations
Abstract
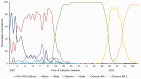
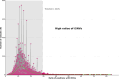
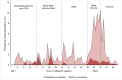
Due to the continuous genetic diversification of severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) over time, the co-circulation of two different lineages in the same region may lead to co-infections within a host, a situation known to contribute to the emergence of hybrid viral populations through genomic recombination. The aim of this study was to use a genomics-based approach to identify distinct viral populations of SARS-CoV-2 in patients with coronavirus disease 2019 (COVID-19), as an indicator of potential co-infections and recombination events. The cohort included 41,224 serial nasopharyngeal swabs positive for SARS-CoV-2 RNA, prospectively collected between January 2021 and April 2022 as part of the French national surveillance program. Full-length genomes were sequenced by next-generation sequencing (COVIDseq). Intra-host single nucleotide variants (iSNVs) were identified, and a synthetic cohort was generated to establish thresholds of co-infection detection. Eight hundred sixty-one samples with iSNV ratios above the threshold were considered "potential co-infections." Peaks in co-infection prevalence occurred during the periods of co-circulation of different SARS-CoV-2 variants. Co-infection with different Variants of Concern (VoC) was confirmed in 103 cases, including Alpha-Beta in 12 cases, Alpha-Delta in 15 cases, Gamma-Delta in 4 cases, Delta-Omicron in 35 cases, and Omicron BA.1-BA.2 in 37 cases. In conclusion, our study suggests a higher prevalence of SARS-CoV-2 variant/subvariant co-infection events than that previously reported using conventional approaches, particularly during periods characterized by the emergence and co-circulation of multiple lineages, creating an increased risk of recombination. Our results support the premise of the importance of genomics-based approaches to detect co-infection events in virus-infected populations, including co-infection with closely related lineages.
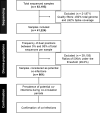
Importance: We aim to implement an innovative approach to monitor and study the diversity of severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) within the human population, particularly during periods of emergence and circulation of VOCs. This approach focused on detecting highly diverse viral samples and co-infection cases, which are known to facilitate viral diversity through recombination and can potentially lead to the emergence of new recombinant lineages with novel characteristics. Monitoring and characterizing co-infection cases during an outbreak is a key strategy for better understanding viral evolution, especially during epidemic periods. However, detecting co-infection cases is challenging, and their prevalence is often highly underestimated. In this study, we developed a strategy to identify highly diverse viral samples that can be implemented in surveillance programs and applied to large datasets. We aim to implement an innovative approach to monitor and study the diversity of SARS-CoV-2 within the human population, particularly during periods of emergence and circulation of Variants of Concern. This approach focused on detecting highly diverse viral samples and co-infection cases, which are known to facilitate viral diversity through recombination and can potentially lead to the emergence of new recombinant lineages with novel characteristics.
Keywords: SARS-CoV-2; co-infections; next-generation sequencing; viral genomic surveillance.
Conflict of interest statement
J.-M.P. has served as an advisor and/or speaker for Abbott, AbbVie, Gilead, and GSK. C.R. has served as a speaker for Pfizer. S.F. has received grants from Moderna and served as a speaker for GlaxoSmithKline, AstraZeneca, MSD, Pfizer, Cepheid, and Moderna. All other authors declare no competing interests.
Figures

References
-
- Kaku Y, Okumura K, Padilla-Blanco M, Kosugi Y, Uriu K, Hinay AA Jr, Chen L, Plianchaisuk A, Kobiyama K, Ishii KJ, Zahradnik J, Ito J, Sato K, Genotype to Phenotype Japan (G2P-Japan) Consortium . 2024. Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect Dis 24:e82. doi:10.1016/S1473-3099(23)00813-7 - DOI - PubMed
-
- Carabelli AM, Peacock TP, Thorne LG, Harvey WT, Hughes J, Peacock SJ, Barclay WS, de Silva TI, Towers GJ, Robertson DL, COVID-19 Genomics UK Consortium . 2023. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol 21:162–177. doi:10.1038/s41579-022-00841-7 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

