Novel endornaviruses infecting Phytophthora cactorum that attenuate vegetative growth, promote sporangia formation and confer hypervirulence to the host oomycete
- PMID: 40310668
- PMCID: PMC12046096
- DOI: 10.1099/jgv.0.002099
Novel endornaviruses infecting Phytophthora cactorum that attenuate vegetative growth, promote sporangia formation and confer hypervirulence to the host oomycete
Abstract
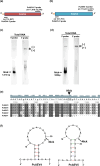
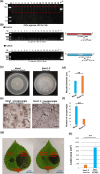
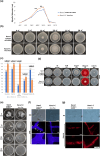
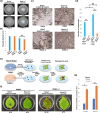
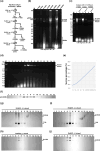
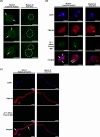
Two novel endornaviruses were found in Phytophthora cactorum isolated from black lesions on Boehmeria nivea var. nipononivea plants in a Japanese forest. These two endornaviruses were named Phytophthora cactorum alphaendornavirus 4 (PcAEV4) and Phytophthora cactorum alphaendornavirus 5 (PcAEV5) and have site-specific nick structures in their positive RNA strands, which are hallmarks of alphaendornaviruses. Ribavirin and cycloheximide treatment of the protoplasts effectively cured the host oomycete (strain Kara1) of the viruses. The resultant virus-free strain (Kara1-C) displayed abundant mycelial growth with less zoosporangia formation as compared to the Kara1 strain. Remarkably, the Kara1-C strain exhibited a reduced ability to form black lesions on B. nivea leaves, suggesting that the presence of PcAEV4 and PcAEV5 in the Kara1 strain led to enhanced virulence in host plants. Under osmotic pressure and cell wall synthesis inhibition, the Kara1 strain exhibited less growth inhibition compared with the Kara1-C strain. In contrast, the Kara1 strain showed more growth inhibition in the presence of membrane-permeable surfactant compared with the Kara1-C strain, indicating that the two endornaviruses can alter the susceptibility of the host oomycete to abiotic stresses. Co-localization and cell fractionation analyses showed that PcAEV4 and PcAEV5 localized to intracellular membranes, particularly the endoplasmic reticulum membrane fraction. Furthermore, infection with these two endornaviruses was found to affect the host's response to exogenous sterols, which enhanced vegetative growth and zoosporangia formation, as well as virulence of the host oomycete. These results provide insights into the effects of endornavirus infection in Phytophthora spp. and also highlight the usefulness of protoplast-based methods in advancing Phytophthora virus studies.
Keywords: Phytophthora; attenuation; endornavirus; hypervirulence; membrane fraction; site-specific nick.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

Similar articles
-
Two Novel Endornaviruses Co-infecting a Phytophthora Pathogen of Asparagus officinalis Modulate the Developmental Stages and Fungicide Sensitivities of the Host Oomycete.Front Microbiol. 2021 Feb 3;12:633502. doi: 10.3389/fmicb.2021.633502. eCollection 2021. Front Microbiol. 2021. PMID: 33633714 Free PMC article.
-
Sequence and phylogenetic analysis of a novel alphaendornavirus, the first virus described from the oomycete plant pathogen Phytophthora heveae.Arch Virol. 2023 May 11;168(6):158. doi: 10.1007/s00705-023-05786-7. Arch Virol. 2023. PMID: 37166518 Free PMC article.
-
Incidence of endornaviruses in Phytophthora taxon douglasfir and Phytophthora ramorum.Virus Genes. 2010 Feb;40(1):130-4. doi: 10.1007/s11262-009-0421-7. Epub 2009 Nov 14. Virus Genes. 2010. PMID: 19915969
-
Endornaviruses: persistent dsRNA viruses with symbiotic properties in diverse eukaryotes.Virus Genes. 2019 Apr;55(2):165-173. doi: 10.1007/s11262-019-01635-5. Epub 2019 Jan 14. Virus Genes. 2019. PMID: 30644058 Review.
-
[Unique symbiotic viruses in plants: Endornaviruses].Uirusu. 2015;65(2):209-218. doi: 10.2222/jsv.65.209. Uirusu. 2015. PMID: 27760919 Review. Japanese.
References
-
- Hollings M. Viruses associated with a die-back disease of cultivated mushroom. Nature. 1962;196:962–965. doi: 10.1038/196962a0. - DOI
-
- Honkura R, Shirako Y, Ehara Y, Yamanaka S. Two types of virus-like particles isolated from downy mildew diseased rice plants. Jpn J Phytopathol. 1983;49:653–658. doi: 10.3186/jjphytopath.49.653. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

