Tebentafusp elicits on-target cutaneous immune responses driven by cytotoxic T cells in uveal melanoma patients
- PMID: 40311102
- PMCID: PMC12165791
- DOI: 10.1172/JCI181464
Tebentafusp elicits on-target cutaneous immune responses driven by cytotoxic T cells in uveal melanoma patients
Abstract
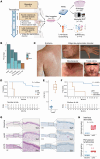
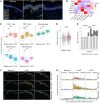
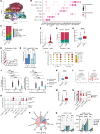
BACKGROUNDTebentafusp is the first T cell receptor-based bispecific protein approved for clinical use in HLA-A*02:01+ adult patients with unresectable/metastatic uveal melanoma. It redirects T cells toward gp100-expressing target cells, frequently inducing skin-related early adverse events.METHODSThis study investigated immunological and cellular responses using single-cell and spatial analysis of skin biopsies from patients with metastatic uveal melanoma treated with tebentafusp.RESULTS81.8% of patients developed acute cutaneous adverse events, which correlated with improved survival. Multimodal analysis revealed a brisk infiltration of CD4+ and CD8+ T cells, while melanocyte numbers declined. Single-cell RNA-sequencing revealed T cell activation, proliferation, and IFN-γ/cytotoxic gene upregulation. CD8+ T cells colocalized with melanocytes and upregulated LAG3, suggesting potential for combination therapies with tebentafusp. Melanocytes upregulated antigen presentation and apoptotic pathways, while pigmentation gene expression decreased. However, gp100 remained stably expressed.CONCLUSIONSequential skin biopsies enable in vivo pharmacodynamic modeling of tebentafusp, offering insights into immune activation, toxicity, and treatment response. Examining the on-target effects of bispecifics in tissues amenable to longitudinal sampling enhances our understanding of toxicity and therapeutic escape mechanisms, guiding strategies for treatment optimization.FUNDINGCancer Research Foundation, Swiss National Science Foundation (323630_207029, 733 310030_170320, 310030_188450, CRSII5_183478), Iten-Kohaut Foundation, European Research Council no. 882424, University Priority Project Translational Cancer Research of the University of Zurich (UZH), UZH PostDoc grant (K-85810-02-01).
Keywords: Cancer immunotherapy; Dermatology; Melanoma; Oncology; Skin.
Figures


References
-
- Cole D, et al. Abstract 2271: Tebentafusp recognition of melanoma cells is restricted by HLA-A0201 presentation of a gp100 peptide. Cancer Res. 2020;80(16_suppl):2271
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

