Single-cell transcriptomic analysis reveals gut microbiota-immunotherapy synergy through modulating tumor microenvironment
- PMID: 40312419
- PMCID: PMC12045981
- DOI: 10.1038/s41392-025-02226-7
Single-cell transcriptomic analysis reveals gut microbiota-immunotherapy synergy through modulating tumor microenvironment
Abstract
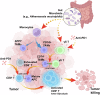
The gut microbiota crucially regulates the efficacy of immune checkpoint inhibitor (ICI) based immunotherapy, but the underlying mechanisms remain unclear at the single-cell resolution. Using single-cell RNA sequencing and subsequent validations, we investigate gut microbiota-ICI synergy by profiling the tumor microenvironment (TME) and elucidating critical cellular interactions in mouse models. Our findings reveal that intact gut microbiota combined with ICIs may synergistically increase the proportions of CD8+, CD4+, and γδ T cells, reduce glycolysis metabolism, and reverse exhausted CD8+ T cells into memory/effector CD8+ T cells, enhancing antitumor response. This synergistic effect also induces macrophage reprogramming from M2 protumor Spp1+ tumor-associated macrophages (TAMs) to Cd74+ TAMs, which act as antigen-presenting cells (APCs). These macrophage subtypes show a negative correlation within tumors, particularly during fecal microbiota transplantation. Depleting Spp1+ TAMs in Spp1 conditional knockout mice boosts ICI efficacy and T cell infiltration, regardless of gut microbiota status, suggesting a potential upstream role of the gut microbiota and highlighting the crucial negative impact of Spp1+ TAMs during macrophage reprogramming on immunotherapy outcomes. Mechanistically, we propose a γδ T cell-APC-CD8+ T cell axis, where gut microbiota and ICIs enhance Cd40lg expression on γδ T cells, activating Cd40 overexpressing APCs (e.g., Cd74+ TAMs) through CD40-CD40L-related NF-κB signaling and boosting CD8+ T cell responses via CD86-CD28 interactions. These findings highlight the potential importance of γδ T cells and SPP1-related macrophage reprogramming in activating CD8+ T cells, as well as the synergistic effect of gut microbiota and ICIs in immunotherapy through modulating the TME.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics: Experiments were conducted in compliance with all relevant governmental and institutional guidelines and regulations. All animal procedures were approved by the Institutional Animal Care and Use Committee of West China Hospital, Sichuan University (Approval No. 20220301038).
Figures







References
-
- Kim, J. M. & Chen, D. S. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann. Oncol.27, 1492–1504 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

