Sex-differentiated placental methylation and gene expression regulation has implications for neonatal traits and adult diseases
- PMID: 40312437
- PMCID: PMC12045980
- DOI: 10.1038/s41467-025-58128-3
Sex-differentiated placental methylation and gene expression regulation has implications for neonatal traits and adult diseases
Abstract
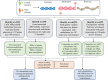
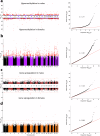
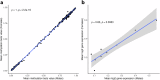
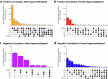
Sex differences in physiological and disease traits are pervasive and begin during early development, but the genetic architecture of these differences is largely unknown. Here, we leverage the human placenta, a transient organ during pregnancy critical to fetal development, to investigate the impact of sex in the regulatory landscape of placental autosomal methylome and transcriptome, and its relevance to health and disease. We find that placental methylation and its genetic regulation are extensively impacted by fetal sex, whereas sex differences in placental gene expression and its genetic regulation are limited. We identify molecular processes and regulatory targets that are enriched in a sex-specific manner, and find enrichment of imprinted genes in sex-differentiated placental methylation, including female-biased methylation within the well-known KCNQ1OT1/CDKN1C imprinting cluster of genes expressed in a parent-of-origin dependent manner. We establish that several sex-differentiated genetic effects on placental methylation and gene expression colocalize with birthweight and adult disease genetic associations, facilitating mechanistic insights on early life origins of health and disease outcomes shaped by sex.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests. Clinical trial registration: ClinicalTrials.gov, NCT00912132.
Figures




References
-
- Khramtsova, E. A., Davis, L. K. & Stranger, B. E. The role of sex in the genomics of human complex traits. Nat. Rev. Genet20, 173–190 (2019). - PubMed
-
- Clifton, V. L. Review: Sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta31, S33–S39 (2010). - PubMed
-
- Sandovici, I., Fernandez-Twinn, D. S., Hufnagel, A., Constância, M. & Ozanne, S. E. Sex differences in the intergenerational inheritance of metabolic traits. Nat. Metab.4, 507–523 (2022). - PubMed
MeSH terms
Grants and funding
- HHSN275200800014C/HD/NICHD NIH HHS/United States
- HHSN275200800002C/HD/NICHD NIH HHS/United States
- HHSN275200800013C/HD/NICHD NIH HHS/United States
- U24 ES028507/ES/NIEHS NIH HHS/United States
- HHSN275200800003I/HD/NICHD NIH HHS/United States
- Intramural Research Program/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- R01 HD108310/HD/NICHD NIH HHS/United States
- HHSN275200800003C/HD/NICHD NIH HHS/United States
- HHSN275201000009C/HD/NICHD NIH HHS/United States
- HHSN275200800002I/HD/NICHD NIH HHS/United States
- HHSN275200800028C/HD/NICHD NIH HHS/United States
- R01 ES022223/ES/NIEHS NIH HHS/United States
- HHSN275200800012C/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources

