Linking peri-implantitis to microbiome changes in affected implants, healthy implants, and saliva: a cross-sectional pilot study
- PMID: 40313461
- PMCID: PMC12043654
- DOI: 10.3389/fcimb.2025.1543100
Linking peri-implantitis to microbiome changes in affected implants, healthy implants, and saliva: a cross-sectional pilot study
Abstract
Introduction: The rising use of dental implants is accompanied by an expected increase in peri-implant diseases, particularly peri-implantitis (PI), which poses a significant threat to implant success and necessitates a thorough understanding of its pathogenesis for effective management.
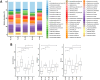
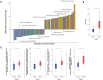
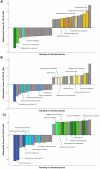
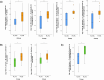
Methods: To gain deeper insights into the role and impact of the peri-implant microbiome in the pathogenesis and progression of PI, we analyzed 100 samples of saliva and subgingival biofilm from 40 participants with healthy implants (HI group) or with co-occurrence of diagnosed PI-affected implants and healthy implants (PI group) using shotgun metagenomic sequencing. We identified the most discriminative species distinguishing healthy from diseased study groups through log ratios and differential ranking analyses.
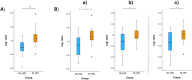
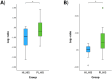
Results and discussion: Mogibacterium timidum, Schaalia cardiffensis, Parvimonas micra, Filifactor alocis, Porphyromonas endodontalis, Porphyromonas gingivalis and Olsenella uli were associated with the subgingival peri-implant biofilm. In contrast, Neisseria sp oral taxon 014, Haemophilus parainfluenzae, Actinomyces naeslundii, Rothia mucilaginosa and Rothia aeria were more prevalent in the healthy peri-implant biofilm. Functional pathways such as arginine and polyamine biosynthesis, including putrescine and citrulline biosynthesis, showed stronger correlations with PI-affected implants. In contrast, peri-implant health was characterized by the predominance of pathways involved in purine and pyrimidine deoxyribonucleotide de novo biosynthesis, glucose and glucose-1-phosphate degradation, and tetrapyrrole biosynthesis. Our findings reveal that healthy implants in PI-free oral cavities differ significantly in microbial composition and functional pathways compared to healthy implants co-occurring with PI-affected implants, which more closely resemble PI-associated profiles. This pattern extended to salivary samples, where microbial and functional biomarkers follow similar trends.
Keywords: compositional change; differential rankings; functional pathways; peri-implant microbiome; peri-implantitis; saliva microbiome; shotgun metagenomic sequencing.
Copyright © 2025 Bessa, Egas, Pires, Proença, Mascarenhas, Pais, Barroso, Machado, Botelho, Alcoforado, Mendes and Alves.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

