Discovery, design, and engineering of enzymes based on molecular retrobiosynthesis
- PMID: 40313979
- PMCID: PMC12042125
- DOI: 10.1002/mlf2.70009
Discovery, design, and engineering of enzymes based on molecular retrobiosynthesis
Abstract
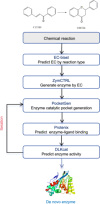
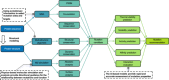
Biosynthesis-a process utilizing biological systems to synthesize chemical compounds-has emerged as a revolutionary solution to 21st-century challenges due to its environmental sustainability, scalability, and high stereoselectivity and regioselectivity. Recent advancements in artificial intelligence (AI) are accelerating biosynthesis by enabling intelligent design, construction, and optimization of enzymatic reactions and biological systems. We first introduce the molecular retrosynthesis route planning in biochemical pathway design, including single-step retrosynthesis algorithms and AI-based chemical retrosynthesis route design tools. We highlight the advantages and challenges of large language models in addressing the sparsity of chemical data. Furthermore, we review enzyme discovery methods based on sequence and structure alignment techniques. Breakthroughs in AI-based structural prediction methods are expected to significantly improve the accuracy of enzyme discovery. We also summarize methods for de novo enzyme generation for nonnatural or orphan reactions, focusing on AI-based enzyme functional annotation and enzyme discovery techniques based on reaction or small molecule similarity. Turning to enzyme engineering, we discuss strategies to improve enzyme thermostability, solubility, and activity, as well as the applications of AI in these fields. The shift from traditional experiment-driven models to data-driven and computationally driven intelligent models is already underway. Finally, we present potential challenges and provide a perspective on future research directions. We envision expanded applications of biocatalysis in drug development, green chemistry, and complex molecule synthesis.
Keywords: artificial intelligence; enzyme design; enzyme discovery; enzyme engineering; molecular retrosynthesis planning.
© 2025 The Author(s). mLife published by John Wiley & Sons Australia, Ltd on behalf of Institute of Microbiology, Chinese Academy of Sciences.
Figures

Similar articles
-
Artificial Intelligence in Retrosynthesis Prediction and its Applications in Medicinal Chemistry.J Med Chem. 2025 Feb 13;68(3):2333-2355. doi: 10.1021/acs.jmedchem.4c02749. Epub 2025 Jan 30. J Med Chem. 2025. PMID: 39883477 Review.
-
Artificial Intelligence Methods and Models for Retro-Biosynthesis: A Scoping Review.ACS Synth Biol. 2024 Aug 16;13(8):2276-2294. doi: 10.1021/acssynbio.4c00091. Epub 2024 Jul 24. ACS Synth Biol. 2024. PMID: 39047143 Free PMC article.
-
AI-driven de novo enzyme design: Strategies, applications, and future prospects.Biotechnol Adv. 2025 Sep;82:108603. doi: 10.1016/j.biotechadv.2025.108603. Epub 2025 May 12. Biotechnol Adv. 2025. PMID: 40368118 Review.
-
Hybrid schemes based on quantum mechanics/molecular mechanics simulations goals to success, problems, and perspectives.Adv Protein Chem Struct Biol. 2011;85:81-142. doi: 10.1016/B978-0-12-386485-7.00003-X. Adv Protein Chem Struct Biol. 2011. PMID: 21920322 Review.
-
Reaction Templates: Bridging Synthesis Knowledge and Artificial Intelligence.Acc Chem Res. 2024 Jul 16;57(14):1964-1972. doi: 10.1021/acs.accounts.4c00261. Epub 2024 Jun 26. Acc Chem Res. 2024. PMID: 38924502
References
-
- Rahman FA, Aziz MMA, Saidur R, Bakar WAWA, Hainin MR, Putrajaya R, et al. Pollution to solution: capture and sequestration of carbon dioxide (CO2) and its utilization as a renewable energy source for a sustainable future. Renew Sust Energy Rev. 2017;71:112–126.
-
- Hao C, Xu L, Kuang H, Xu C. Artificial chiral probes and bioapplications. Adv Mater. 2020;32:e1802075. - PubMed
-
- Sharma A, Gupta G, Ahmad T, Mansoor S, Kaur B. Enzyme engineering: current trends and future perspectives. Food Rev Int. 2021;37:121–154.
Publication types
LinkOut - more resources
Full Text Sources
