[18F]flutemetamol uptake in the colon of a memory clinic population and its association with brain amyloidosis and the gut microbiota profile: an exploratory study
- PMID: 40314812
- PMCID: PMC12491377
- DOI: 10.1007/s00259-025-07299-8
[18F]flutemetamol uptake in the colon of a memory clinic population and its association with brain amyloidosis and the gut microbiota profile: an exploratory study
Abstract
Purpose: Some Alzheimer's disease (AD) patients report gastro-intestinal symptoms and present alterations in the gut microbiota (GM) composition. Elevated colonic amyloid immunoreactivity has been shown in patients and animal models. We evaluated the colonic uptake of the amyloid positron emission tomography (PET) imaging agent [18F]flutemetamol (FMM) in a memory clinic population and investigated its association with brain amyloidosis and GM composition.
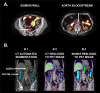
Methods: Forty-five participants underwent (i) abdominal and cerebral FMM PET, acquired at 40 (early phase) and 120 min (late phase) after tracer injection, (ii) abdominal computed tomography, and (iii) cerebral T1-weighted MRI. Colonic standardized uptake value ratio (SUVr) was determined through manual tracing and automatic segmentation (TotalSegmentator), using the aortic blood signal as a reference region. Fecal GM composition was assessed using 16 S rRNA sequencing. Amyloid positive (A+) and negative (A-) participants, based on cortical FMM quantification (PetSurfer), were compared in terms of SUVr and GM features.
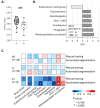
Results: Increased colonic early SUVr was reported in A+ than A- (manual, p =.008; automated, p =.035). Altered GM composition was found in A + as shown by lower Pielou's evenness (p =.023), lower abundance of Eubacterium hallii group, and higher abundance of several genera. High UC5-1-2E3 abundance positively correlated with high colonic early SUVr (whole group: manual, p =.012, automated, p =.082; A+: manual, p =.074; automated, p =.016).
Conclusion: This exploratory study showed that subjects with cerebral amyloidosis have greater colonic FMM uptake than subjects with normal cerebral amyloid load, correlating with altered GM composition. Further analysis is needed to determine if these changes denote amyloid-related changes or other phenomena.
Keywords: Alzheimer’s disease; Amyloid PET; Colon; Gut microbiota.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: The gMAD/COSCODE study was approved by the competent ethics committee (Commission cantonale d’éthique de la recherche [CCER]; reference No. CCER_2016 − 01346, CCER_2020_00403) and was conducted in accordance with the first revision of the Declaration of Helsinki from 1975. Consent to participate: Informed consent was obtained from all individual participants included in the study. Competing interests: The authors declare that they have no competing interests.
Figures

References
-
- Scheltens P, Blennow K, Breteler MMB, de Strooper B, Frisoni GB, Salloway S, et al. Alzheimer’s disease. Lancet. 2016;388:505–17. 10.1016/S0140-6736(15)01124-1 - PubMed
-
- Hernández-Ruiz V, Roubaud-Baudron C, Von Campe H, Retuerto N, Mégraud F, Helmer C, et al. Association between Helicobacter pylori infection and incident risk of dementia: the AMI cohort. J Am Geriatr Soc. 2024;72:1191–8. 10.1111/jgs.18748 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

