Accurate, scalable, and fully automated inference of species trees from raw genome assemblies using ROADIES
- PMID: 40314967
- PMCID: PMC12088440
- DOI: 10.1073/pnas.2500553122
Accurate, scalable, and fully automated inference of species trees from raw genome assemblies using ROADIES
Abstract
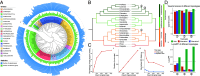
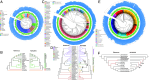
Current genome sequencing initiatives across a wide range of life forms offer significant potential to enhance our understanding of evolutionary relationships and support transformative biological and medical applications. Species trees play a central role in many of these applications; however, despite the widespread availability of genome assemblies, accurate inference of species trees remains challenging due to the limited automation, substantial domain expertise, and computational resources required by conventional methods. To address this limitation, we present ROADIES, a fully automated pipeline to infer species trees starting from raw genome assemblies. In contrast to the prominent approach, ROADIES incorporates a unique strategy of randomly sampling segments of the input genomes to generate gene trees. This eliminates the need for predefining a set of loci, limiting the analyses to a fixed number of genes, and performing the cumbersome gene annotation and/or whole genome alignment steps. ROADIES also eliminates the need to infer orthology by leveraging existing discordance-aware methods that allow multicopy genes. Using the genomic datasets from large-scale sequencing efforts across four diverse life forms (placental mammals, pomace flies, birds, and budding yeasts), we show that ROADIES infers species trees that are comparable in quality to the state-of-the-art studies but in a fraction of the time and effort, including on challenging datasets with rampant gene tree discordance and complex polyploidy. With its speed, accuracy, and automation, ROADIES has the potential to vastly simplify species tree inference, making it accessible to a broader range of scientists and applications.
Keywords: bioinformatics; phylogenetics; species tree inference.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

Update of
-
Accurate, scalable, and fully automated inference of species trees from raw genome assemblies using ROADIES.bioRxiv [Preprint]. 2024 Jun 1:2024.05.27.596098. doi: 10.1101/2024.05.27.596098. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2025 May 13;122(19):e2500553122. doi: 10.1073/pnas.2500553122. PMID: 38854139 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

