Glycolytic disruption restricts Drosophila melanogaster larval growth via the cytokine Upd3
- PMID: 40315265
- PMCID: PMC12068724
- DOI: 10.1371/journal.pgen.1011690
Glycolytic disruption restricts Drosophila melanogaster larval growth via the cytokine Upd3
Abstract
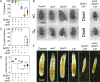
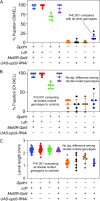
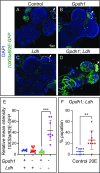
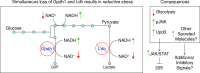
Drosophila larval growth requires efficient conversion of dietary nutrients into biomass. Lactate dehydrogenase (Ldh) and glycerol-3-phosphate dehydrogenase (Gpdh1) support this larval metabolic program by cooperatively promoting glycolytic flux. Consistent with their cooperative functions, the loss of both enzymes, but not either single enzyme alone, induces a developmental arrest. However, Ldh and Gpdh1 exhibit complex and often mutually exclusive expression patterns, suggesting that the lethal phenotypes exhibited by Gpdh1; Ldh double mutants could be mediated non-autonomously. Supporting this possibility, we find that the developmental arrest displayed by double mutants extends beyond simple metabolic disruption and instead stems, in part, from changes in systemic growth factor signaling. Specifically, we demonstrate that the simultaneous loss of Gpdh1 and Ldh results in elevated expression of Upd3, a cytokine involved in Jak/Stat signaling. Furthermore, we show that upd3 loss-of-function mutations suppress the Gpdh1; Ldh larval arrest phenotype, indicating that Upd3 signaling restricts larval development in response to decreased glycolytic flux. Together, our findings reveal a mechanism by which metabolic disruptions can modulate systemic growth factor signaling.
Copyright: © 2025 Rai et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
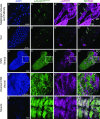
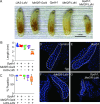
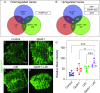
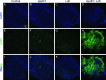

Similar articles
-
Glycolytic Disruption Triggers Interorgan Signaling to Nonautonomously Restrict Drosophila Larval Growth.bioRxiv [Preprint]. 2024 Jun 9:2024.06.06.597835. doi: 10.1101/2024.06.06.597835. bioRxiv. 2024. PMID: 38895259 Free PMC article. Preprint.
-
Lactate dehydrogenase and glycerol-3-phosphate dehydrogenase cooperatively regulate growth and carbohydrate metabolism during Drosophila melanogaster larval development.Development. 2019 Sep 12;146(17):dev175315. doi: 10.1242/dev.175315. Development. 2019. PMID: 31399469 Free PMC article.
-
Hypoxia-inducible factor 1α is required to establish the larval glycolytic program in Drosophila melanogaster.Mol Metab. 2025 Mar;93:102106. doi: 10.1016/j.molmet.2025.102106. Epub 2025 Jan 31. Mol Metab. 2025. PMID: 39894213 Free PMC article.
-
The Drosophila melanogaster enzyme glycerol-3-phosphate dehydrogenase 1 is required for oogenesis, embryonic development, and amino acid homeostasis.G3 (Bethesda). 2022 Jul 29;12(8):jkac115. doi: 10.1093/g3journal/jkac115. G3 (Bethesda). 2022. PMID: 35536221 Free PMC article.
-
FLPing Genes On and Off in Drosophila.Methods Mol Biol. 2017;1642:195-209. doi: 10.1007/978-1-4939-7169-5_13. Methods Mol Biol. 2017. PMID: 28815502 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

