Fibroblastic reticular cells form reactive myeloid cell niches in human lymph nodes
- PMID: 40315298
- PMCID: PMC12646167
- DOI: 10.1126/sciimmunol.ads6820
Fibroblastic reticular cells form reactive myeloid cell niches in human lymph nodes
Abstract
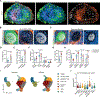
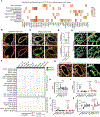
Lymph nodes play a key role in maintaining fluid balance in homeostatic and inflamed tissues and provide fibroblastic niche environments for optimal immune cell positioning and interaction. Here, we used single-cell and spatial transcriptomic analyses in combination with high-resolution imaging to molecularly define and functionally characterize niche-forming cells that control inflammation-driven remodeling in human lymph nodes. Fibroblastic reticular cells responded to inflammatory perturbation with activation and expansion of poised niche environments. Inflammation-induced adaptation of lymph node infrastructure and topography included the expansion of peptidase inhibitor 16 (PI16)-expressing reticular cell (PI16+ RC) networks that enwrap the perivenular conduit system. Interactome analyses indicated that macrophage-derived oncostatin M directs PI16+ RC activation in inflamed lymph nodes and thereby promotes immune cell aggregation in the perivenular space. In conclusion, these data demonstrate that the inflammatory remodeling of human lymph nodes results in the formation of reactive myeloid cell niches by PI16+ RCs.
Conflict of interest statement
Competing interests
H.-W.C., L.O., N.B.P. and B.L. are founders and H.-W.C., L.O., N.B.P., and B.L. are shareholders of Stromal Therapeutics AG, Basel, Switzerland. L.O. and B.L. are members of the board of Stromal Therapeutics AG, Basel, Switzerland. L.O. and B.L. are listed as inventors on patent WO 2022/084400 A1. I.M. has received research funding from Genentech and Regeneron, and is a member of Garuda Therapeutics’s scientific advisory board. All other authors declare no competing interests.
Figures





References
-
- Meizlish ML, Franklin RA, Zhou X, Medzhitov R, Tissue Homeostasis and Inflammation. Annu Rev Immunol 39, 557–581 (2021). - PubMed
-
- Onder L, Ludewig B, A Fresh View on Lymph Node Organogenesis. Trends Immunol 39, 775–787 (2018). - PubMed
-
- Acton SE, Onder L, Novkovic M, Martinez VG, Ludewig B, Communication, construction, and fluid control: lymphoid organ fibroblastic reticular cell and conduit networks. Trends Immunol 42, 782–794 (2021). - PubMed
-
- Boehm T, Hess I, Swann JB, Evolution of lymphoid tissues. Trends Immunol 33, 315–321 (2012). - PubMed
-
- Junt T, Scandella E, Ludewig B, Form follows function: lymphoid tissue microarchitecture in antimicrobial immune defence. Nat. Rev. Immunol 8, 764–775 (2008). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

