Noninvasive prognostic classification of ITH in HCC with multi-omics insights and therapeutic implications
- PMID: 40315307
- PMCID: PMC12047409
- DOI: 10.1126/sciadv.ads8323
Noninvasive prognostic classification of ITH in HCC with multi-omics insights and therapeutic implications
Abstract
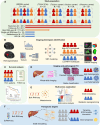
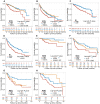
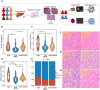
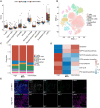
Intratumoral heterogeneity (ITH) is a critical factor associated with treatment failure and disease relapse in hepatocellular carcinoma (HCC). However, decoding ITH in a noninvasive and comprehensive manner remains a notable challenge. In this study involving 851 patients from five centers, we developed a noninvasive prognostic classification for ITH using radiomics based on multisequence MRI, termed radiomics ITH (RITH) phenotypes. The RITH phenotypes highly correlated with prognosis and pathological ITH. In addition, through an integrated multi-omics analysis, we uncovered the molecular mechanisms underlying RITH, notably enhancing its biological interpretability. Specifically, high-RITH tumors demonstrated an enrichment of cancer-associated fibroblasts and activation of extracellular matrix remodeling. Our approach facilitates the noninvasive refined classification of ITH using radiomics and multi-omics, paving the way for tailored treatment strategies in HCC. Extracellular matrix-receptor interaction could be a potential therapeutic target in patients with high-RITH tumors. Given the routine use of radiologic imaging in oncology, our methodology ignites versatile framework for broader application to other solid tumors.
Figures



References
-
- Dentro S. C., Leshchiner I., Haase K., Tarabichi M., Wintersinger J., Deshwar A. G., Yu K., Rubanova Y., Macintyre G., Demeulemeester J., Vázquez-García I., Kleinheinz K., Livitz D. G., Malikic S., Donmez N., Sengupta S., Anur P., Jolly C., Cmero M., Rosebrock D., Schumacher S. E., Fan Y., Fittall M., Drews R. M., Yao X., Watkins T. B. K., Lee J., Schlesner M., Zhu H., Adams D. J., McGranahan N., Swanton C., Getz G., Boutros P. C., Imielinski M., Beroukhim R., Sahinalp S. C., Ji Y., Peifer M., Martincorena I., Markowetz F., Mustonen V., Yuan K., Gerstung M., Spellman P. T., Wang W., Morris Q. D., Wedge D. C., Van Loo P., PCAWG Evolution and Heterogeneity Working Group and the PCAWG Consortium , Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell 184, 2239–2254.e39 (2021). - PMC - PubMed
-
- McGranahan N., Swanton C., Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 168, 613–628 (2017). - PubMed
-
- An F. Q., Matsuda M., Fujii H., Tang R. F., Amemiya H., Dai Y. M., Matsumoto Y., Tumor heterogeneity in small hepatocellular carcinoma: Analysis of tumor cell proliferation, expression and mutation of p53 AND beta-catenin. Int. J. Cancer 93, 468–474 (2001). - PubMed
-
- Kenmochi K., Sugihara S., Kojiro M., Relationship of histologic grade of hepatocellular carcinoma (HCC) to tumor size, and demonstration of tumor cells of multiple different grades in single small HCC. Liver 7, 18–26 (1987). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

