Taxonomically distinct diatom viruses differentially impact microbial processing of organic matter
- PMID: 40315326
- PMCID: PMC12047433
- DOI: 10.1126/sciadv.adq5439
Taxonomically distinct diatom viruses differentially impact microbial processing of organic matter
Abstract
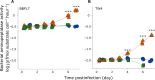
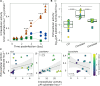
Phytoplankton viruses facilitate the production of dissolved organic matter (DOM) through host lysis, shaping DOM composition, and subsequent regenerative processing. We explored how DOM generated from a bloom-forming, centric diatom, infected with taxonomically distinct viruses-a single-stranded (ss) DNA and a ssRNA virus-impacted microbial processing of organic matter. DOM derived from uninfected and ssDNA virus-infected cultures supported growth in bacterial isolates and a mixed assemblage. In contrast, DOM from ssRNA virus infection did not stimulate growth, but rather induced ectoproteolytic activity, suggesting this DOM was less bioavailable. Exoprotease activity was also substantially higher in ssRNA virus-infected cellular exudates compared to ssDNA virus-infected and uninfected cultures. This suggests that DOM produced through virus-mediated host lysis does not a priori support secondary production and implicate ssRNA virus infection as a source of proteolytic activity in the water column, highlighting a multifaceted role for viruses in altering microbial utilization and remineralization length scales of organic matter in the ocean.
Figures



References
-
- Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P., Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 281, 237–240 (1998). - PubMed
-
- C. A. Carlson, D. A. Hansell, “Chapter 3 - DOM sources, sinks, reactivity, and budgets” in Biogeochemistry of Marine Dissolved Organic Matter (Academic Press, 2014), pp. 65–126.
-
- D. J. Repeta, “Chapter 2 - Chemical characterization and cycling of dissolved organic matter” in Biogeochemistry of Marine Dissolved Organic Matter (Academic Press, 2014), pp. 21–63.
-
- Moran M. A., Kujawinski E. B., Stubbins A., Fatland R., Aluwihare L. I., Buchan A., Crump B. C., Dorrestein P. C., Dyhrman S. T., Hess N. J., Howe B., Longnecker K., Medeiros P. M., Niggemann J., Obernosterer I., Repeta D. J., Waldbauer J. R., Deciphering ocean carbon in a changing world. Proc. Natl. Acad. Sci. U.S.A. 113, 3143–3151 (2016). - PMC - PubMed
-
- Buchan A., LeCleir G. R., Gulvik C. A., González J. M., Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698 (2014). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

