Tumor-infiltrated double-negative regulatory T cells predict outcome of T cell-based immunotherapy in nasopharyngeal carcinoma
- PMID: 40315843
- PMCID: PMC12147895
- DOI: 10.1016/j.xcrm.2025.102096
Tumor-infiltrated double-negative regulatory T cells predict outcome of T cell-based immunotherapy in nasopharyngeal carcinoma
Abstract
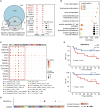
Adoptive cell therapy (ACT) using tumor-infiltrating lymphocytes (TILs) has demonstrated clinical success in solid tumors. We analyze 47 TIL infusion products and 62 pretreatment tumor microenvironments (TMEs) from a randomized phase 2 clinical study of concurrent chemoradiotherapy plus TIL-ACT (NCT02421640). Using single-cell and bulk RNA sequencing along with flow cytometry, we identify 14 CD3+ T cell clusters within 26 TIL infusion products: 11 CD3+CD8+ TILs, 2 CD3+CD4+ TILs, and 1 CD3+CD8-CD4- double-negative (DN) TIL. (DN) TILs, significantly associated with poor TIL-ACT outcomes, exhibit an activated regulatory T cell-like phenotype and include two CD56+ and four CD56- subsets. Among them, CD56-KZF2+ (DN) TILs are predominantly suppressive. (DN) TILs inhibit CD8+ TIL expansion via Fas-FasL, transforming growth factor β (TGF-β), and interleukin (IL)-10 signaling. Distinct CD8+ T subsets differentially impact on TIL-ACT outcomes, while 9 baseline TME gene signatures and 14 intracellular T cell genes hold prognostic value. Our findings identify predictive TIL subsets and biomarkers for TIL-ACT outcomes.
Keywords: TIL infusion products; adoptive cell therapy; double-negative T cells; nasopharyngeal carcinoma; single-cell RNA sequencing.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
-
- Rohaan M.W., Borch T.H., van den Berg J.H., Met Ö., Kessels R., Geukes Foppen M.H., Stoltenborg Granhøj J., Nuijen B., Nijenhuis C., Jedema I., et al. Tumor-Infiltrating Lymphocyte Therapy or Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2022;387:2113–2125. doi: 10.1056/NEJMoa2210233. - DOI - PubMed
-
- Amaria R., Knisely A., Vining D., Kopetz S., Overman M.J., Javle M., Antonoff M.B., Tzeng C.W.D., Wolff R.A., Pant S., et al. Efficacy and safety of autologous tumor-infiltrating lymphocytes in recurrent or refractory ovarian cancer, colorectal cancer, and pancreatic ductal adenocarcinoma. J. Immunother. Cancer. 2024;12 doi: 10.1136/jitc-2023-006822. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

