Exploring prokaryotic diversity in permafrost-affected soils of Ladakh's Changthang region and its geochemical drivers
- PMID: 40316627
- PMCID: PMC12048601
- DOI: 10.1038/s41598-025-94542-9
Exploring prokaryotic diversity in permafrost-affected soils of Ladakh's Changthang region and its geochemical drivers
Abstract
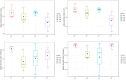
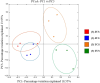
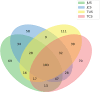
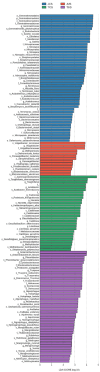
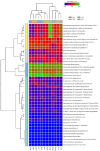
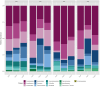
Global warming due to climate change has substantial impact on high-altitude permafrost affected soils. This raises a serious concern that the microbial degradation of sequestered carbon can result in alteration of the biogeochemical cycles. Therefore, the characterization of permafrost affected soil microbiomes, especially of unexplored high-altitude, low oxygen arid region, is important for predicting their response to climate change. This study presents the first report of the bacterial diversity of permafrost-affected soils in the Changthang region of Ladakh. The relationship between soil pH, organic carbon, electrical conductivity, and available micronutrients with the microbial diversity was investigated. Amplicon sequencing of permafrost affected soil samples from Jukti and Tsokar showed that Proteobacteria and Actinobacteria were the dominant phyla in all samples. The genera Brevitalea, Chthoniobacter, Sphingomonas, Hydrogenispora, Clostridium, Gaiella, Gemmatimonas were relatively abundant in the Jukti samples whereas the genera Thiocapsa, Actinotalea, Syntrophotalea, Antracticibcterium, Luteolibacter, Nitrospirillum dominated the Tsokar sample. Correlation analyses highlighted the influence of soil geochemical parameters on the bacterial community structure. PCoA analyses showed that the bacterial beta diversity varied significantly between the sampling locations (PERMANOVA test (F-value: 2.3316; R2 = 0.466, p = 0.001) and similar results were also obtained while comparing genus abundance data using the ANOSIM test (R = 0.345, p = 0.007).
Keywords: 16S rRNA; Alpine permafrost; Bacteria; Illumina sequencing; Microbiome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




References
-
- Van Everdingen, R. O. (ed) Multi-language Glossary of Permafrost and Related ground-ice Terms in Chinese, English, French, German, Icelandic, Italian, Norwegian, Polish, Romanian, Russian, Spanish, and Swedish (International Permafrost Association, Terminology Working Group, 1998).
-
- Margesin, R. Permafrost Soils (Springer, 2009).
-
- Gilichinsky, D. et al. Bacteria in Permafrost Psychrophiles: From Biodiversity to Biotechnology 83–102 (Springer, 2008).
-
- Williams, P. J. & Smith, M. W. The Frozen Earth: Fundamentals of Geocryology (Cambridge University Press, 1989).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

