Cellular parameters shaping pathways of targeted protein degradation
- PMID: 40316744
- PMCID: PMC12048530
- DOI: 10.1038/s42003-025-08104-w
Cellular parameters shaping pathways of targeted protein degradation
Abstract
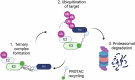
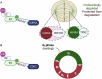
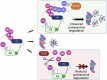
In recent years the development of proteolysis-targeting chimeras (PROTACs) has enhanced the field of ubiquitin signalling through advancing therapeutic targeted protein degradation (TPD) strategies and generating tools to explore the ubiquitin landscape. However, the interplay between PROTACs and their substrates, and other components of the ubiquitin proteasome system (UPS), raises fundamental questions about cellular parameters that might influence the action of PROTACs and the amenability of a given target to PROTAC-mediated degradation. In this perspective we discuss examples of cellular parameters that have been shown to influence PROTAC sensitivity and consider others likely to be important for PROTAC-mediated target degradation but not yet routinely considered in design of novel TPD strategies: Target localisation and accessibility on the one hand, and expression patterns, localisation and activity of E3 ligases, deubiquitinases (DUBs) and wider ubiquitin machinery on the other, are critical parameters in the exploitation of PROTACs, and establishing a better understanding of these parameters will facilitate the rational design of PROTACs.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

