TJ0113-induced mitophagy in acute liver failure detected by Raman microspectroscopy
- PMID: 40318303
- PMCID: PMC12098161
- DOI: 10.1016/j.redox.2025.103654
TJ0113-induced mitophagy in acute liver failure detected by Raman microspectroscopy
Abstract
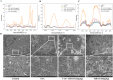
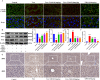
Impaired mitophagy underlies the pathophysiology of acute liver failure (ALF) and is closely associated with tissue damage and dysfunction. A novel mitophagy inducer, TJ0113, was used for treatment during ALF pathogenesis. In this study, we used a novel mitophagy inducer, TJ0113, to investigate the effects and mechanisms of TAA-induced ALF mice. The results showed that TJ0113 could enhance mitophagy through Parkin/PINK1 and ATG5 pathways, which in turn attenuated mitochondrial damage, hepatocyte apoptosis, nuclear factor (NF)-κB/NLRP3 signaling activation and inflammatory responses after TAA. Metabolomics results showed that TJ0113 mainly regulated lipid metabolism, amino acid metabolism and nucleotide metabolism in the livers of ALF mice. RNA sequencing (RNA-seq) analysis yielded that TJ0113 was involved in the development of ALF by regulating the P13K/AKT signaling pathway. The key highlight of this work is the use of an aberration-free line-scanning confocal Raman imager (AFLSCRI) to study the molecular changes in blood, liver tissue, gastrocnemius muscle, and mitochondrial extracts in ALF mice after TJ0113 treatment. Compared to the measurement with conventional assays, Raman microspectroscopy (micro-Raman) offers the benefits of being rapid, non-invasive, label-free and real-time. Our results found good agreement between Raman signals and histopathologic findings. The system has good performance with a spatial resolution of 2 μm, a spectral resolution of 4 cm-1 and a fast detection speed improved by 2 orders. Innovations in this test contribute to clinical diagnosis of disease, personalized treatment, effective intraoperative guidance and accurate prognosis. The data may help in the development of a non-invasive clinical device for mitochondrial damage using bedside micro-Raman.
Keywords: Aberration-free line-scanning confocal Raman imaging; Acute liver failure; Mitophagy; Point-scan Raman imaging; Raman signal; TJ0113.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest This manuscript has not been published or presented elsewhere in part or in entirety and is not under consideration by another journal. Animal experiments were approved by the Animal Experimentation Ethics Committee of Taizhou Hospital, Zhejiang Province (Approval No. tzy-2023216). We have read and understood your journal's policies, and we believe that neither the manuscript nor the study violates any of these. There are no conflicts of interest to declare.
Figures








Similar articles
-
TJ0113 attenuates fibrosis in metabolic dysfunction-associated steatohepatitis by inducing mitophagy.Int Immunopharmacol. 2025 May 27;156:114678. doi: 10.1016/j.intimp.2025.114678. Epub 2025 Apr 18. Int Immunopharmacol. 2025. PMID: 40252468
-
Quercetin inhibits mitophagy-mediated apoptosis and inflammatory response by targeting the PPARγ/PGC-1α/NF-κB axis to improve acute liver failure.Int Immunopharmacol. 2024 Dec 25;143(Pt 2):113444. doi: 10.1016/j.intimp.2024.113444. Epub 2024 Oct 25. Int Immunopharmacol. 2024. PMID: 39454407
-
Endotoxin tolerance ameliorates lipopolysaccharide/D-galactosamine-induced acute liver failure by negative regulation of the NF-κB/NLRP3 and activation of Nrf2/HO-1 via Sitr1.Int Immunopharmacol. 2024 May 10;132:111994. doi: 10.1016/j.intimp.2024.111994. Epub 2024 Apr 5. Int Immunopharmacol. 2024. PMID: 38581992
-
Promotion of mitochondrial energy metabolism during hepatocyte apoptosis in a rat model of acute liver failure.Mol Med Rep. 2015 Oct;12(4):5035-41. doi: 10.3892/mmr.2015.4029. Epub 2015 Jul 2. Mol Med Rep. 2015. PMID: 26135512 Free PMC article.
-
Targeting Pink1-Parkin-mediated mitophagy for treating liver injury.Pharmacol Res. 2015 Dec;102:264-9. doi: 10.1016/j.phrs.2015.09.020. Epub 2015 Oct 24. Pharmacol Res. 2015. PMID: 26655101 Free PMC article. Review.
References
-
- Bernal W., Wendon J. Acute liver failure. N. Engl. J. Med. 2013;369(26):2525–2534. - PubMed
-
- Bernal W., McPhail M.J. Acute liver failure. J. Hepatol. 2021;74(6):1489–1490. - PubMed
-
- Jiao F.Z., Wang Y., Zhang W.B., Zhang H.Y., Chen Q., Shi C.X., Wang L.W., Gong Z.J. Protective role of AGK2 on thioacetamide-induced acute liver failure in mice. Life Sci. 2019;230:68–75. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

