Genetic ancestry and population structure in the All of Us Research Program cohort
- PMID: 40319026
- PMCID: PMC12049439
- DOI: 10.1038/s41467-025-59351-8
Genetic ancestry and population structure in the All of Us Research Program cohort
Abstract
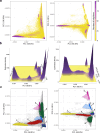
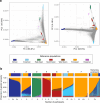
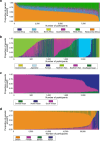
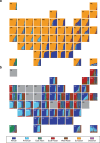
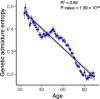
We analyzed participant genomic variant data to characterize population structure and genetic ancestry for the All of Us cohort (n = 297,549). There is substantial population structure in the cohort, with clusters of closely related participants interspersed among less related individuals. Participants show diverse genetic ancestry, with major contributions from European (66.4%), African (19.5%), Asian (7.6%), and American (6.3%) continental ancestry components. Participant genetic similarity clusters show group-specific ancestry, with distinct patterns of continental and subcontinental ancestry among groups. African and American ancestry are enriched in the southeast and southwest regions of the country, respectively, whereas European ancestry is more evenly distributed across the US. The diversity of All of Us participants' genetic ancestry is negatively correlated with age; younger participants show higher levels of genetic admixture compared to older participants. Our results underscore the ancestral genetic diversity of the All of Us cohort, a crucial prerequisite for genomic health equity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
MeSH terms
Grants and funding
- R01 NS112422/NS/NINDS NIH HHS/United States
- OT2 OD026556/OD/NIH HHS/United States
- U24 OD023176/OD/NIH HHS/United States
- 1 OT2 OD025276/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U24 OD023121/OD/NIH HHS/United States
- 1 OT2 OD026554/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U24 OD023176/CD/ODCDC CDC HHS/United States
- OT2 OD025276/OD/NIH HHS/United States
- 1 OT2 OD025277/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- OT2 OD035404/OD/NIH HHS/United States
- 1 OT2 OD35404/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- OT2 OD025315/OD/NIH HHS/United States
- 1 U24 OD023121/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- OT2 OD026551/OD/NIH HHS/United States
- OT2 OD026549/OD/NIH HHS/United States
- 1 ZIAMD000016/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 1 RM1 HG012334/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- OT2 OD025337/OD/NIH HHS/United States
- 1 ZIAMD000018./U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 1 OT2 OD025337/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 1 OT2 OD026557/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- AOD 16037/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- OT2 OD025277/OD/NIH HHS/United States
- RM1 HG012334/HG/NHGRI NIH HHS/United States
- OT2 OD030043/OD/NIH HHS/United States
- 5 R01 NS112422/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 3 OT2 OD025315/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 1 OT2 OD026556/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 1 OT2 OD030043/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- OT2 OD026557/OD/NIH HHS/United States
- 75N98019F01202/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 1 OT2 OD026549/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- OT2 OD026554/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources

