Plasma metabolomics signatures predict COVID-19 patient outcome at ICU admission comparable to clinical scores
- PMID: 40319053
- PMCID: PMC12049461
- DOI: 10.1038/s41598-025-00373-z
Plasma metabolomics signatures predict COVID-19 patient outcome at ICU admission comparable to clinical scores
Abstract
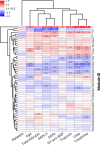
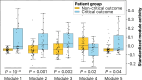
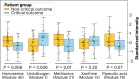
SARS-CoV-2 significantly impacts the human metabolome. This study aims to evaluate the predictive capability of a comprehensive module clustering approach in plasma metabolomics for identifying the risk of critical complications in COVID-19 patients admitted to intensive care units (ICUs). We conducted a prospective monocenter study, gathering blood samples within 24 h of ICU admission, alongside clinical, biological, and demographic patient characteristics. Subsequently, we quantified patients' plasma metabolome using a comprehensive untargeted metabolomics approach. First, we stratified patients based on a composite outcome score indicating critical status. Analysis of potential predictors revealed that older patients with higher severity scores and pronounced alterations in key biological parameters are more likely to experience critical complications. Next, we identified 6,667 metabolic features clustered into 57 annotated metabolic modules across all patients by employing an integrative metabolomics approach. Furthermore, we identified the most differentially expressed metabolic modules related to patients' outcomes. Moreover, we defined the top five most predictive metabolites of critical status: homoserine, urobilinogen, methionine, xanthine and pipecolic acid. These five predictors alone demonstrated similar or superior performance compared to clinical and demographic variables in predicting patients' outcomes. This innovative metabolic module inference approach offers a valuable framework for identifying patients prone to complications upon ICU admission for COVID-19. Its potential applications extend to enhancing patient management across diverse clinical settings.
Keywords: COVID-19; Critical care; Metabolomics; Network clustering; Prediction.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Institutional review board: This study was approved by an independent review board (CPP Est-III, reference 2020-A00885-34) and was registered on https://clinicaltrials.gov/ on April 2020 (reference NCT04357847, principal investigator: Pr E. Besnier). This study was in accordance with the Helsinki Declaration. Informed consent: This study was classified as non-interventional and verbal informed consent was obtained from the patients before their participation.
Figures


References
-
- WHO Coronavirus (COVID-19) dashboard. [cited 22 Jun 2023]. Available: https://covid19.who.int/
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

