Harnessing the FGFR2/NF2/YAP signaling-dependent necroptosis to develop an FGFR2/IL-8 dual blockade therapeutic strategy
- PMID: 40319089
- PMCID: PMC12049493
- DOI: 10.1038/s41467-025-59318-9
Harnessing the FGFR2/NF2/YAP signaling-dependent necroptosis to develop an FGFR2/IL-8 dual blockade therapeutic strategy
Abstract
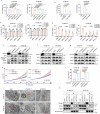
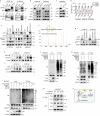
The multifaceted roles and mechanisms of necroptosis in cancer cells remain incompletely understood. Here, we demonstrate that FGFR2 inhibition potently inhibits esophageal squamous cell carcinoma (ESCC) by inducing necroptosis in a RIP1/MLKL-dependent manner and show RIP3 is dispensable in this pathway. Notably, MST1 is identified as a necroptotic pathway component that interacts with RIP1 and MLKL to promote necroptosis by phosphorylating MLKL at Thr216. Additionally, FGFR2 inhibition induces Ser518 phosphorylation and triggers ubiquitin-mediated degradation of NF2, culminating in Hippo pathway suppression. Subsequently, YAP activation promotes RIP1 and MLKL transcriptional upregulation, further amplifying necroptosis. Intriguingly, IL-8 derived from necrotic cells stimulates peripheral surviving tumor cells to increase PD-L1 expression. Dual blockade of FGFR2/PD-L1 or FGFR2/IL-8-CXCR1/2 robustly impedes tumor growth in humanized mouse xenografts. Collectively, our findings delineate an alternative FGFR2-NF2-YAP signaling-dependent necroptotic pathway and shed light on the immunoregulatory role of FGFR2, offering promising avenues for combinatorial therapeutic strategies in clinical cancer management.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

