Alzheimer's disease protective allele of Clusterin modulates neuronal excitability through lipid-droplet-mediated neuron-glia communication
- PMID: 40319306
- PMCID: PMC12049787
- DOI: 10.1186/s13024-025-00840-1
Alzheimer's disease protective allele of Clusterin modulates neuronal excitability through lipid-droplet-mediated neuron-glia communication
Abstract
Background: Genome-wide association studies (GWAS) of Alzheimer's disease (AD) have identified a plethora of risk loci. However, the disease variants/genes and the underlying mechanisms have not been extensively studied.
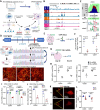
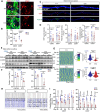
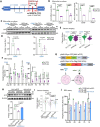
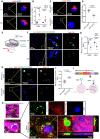
Methods: Bulk ATAC-seq was performed in induced pluripotent stem cells (iPSCs) differentiated various brain cell types to identify allele-specific open chromatin (ASoC) SNPs. CRISPR-Cas9 editing generated isogenic pairs, which were then differentiated into glutamatergic neurons (iGlut). Transcriptomic analysis and functional studies of iGlut co-cultured with mouse astrocytes assessed neuronal excitability and lipid droplet formation.
Results: We identified a putative causal SNP of CLU that impacted neuronal chromatin accessibility to transcription-factor(s), with the AD protective allele upregulating neuronal CLU and promoting neuron excitability. And, neuronal CLU facilitated neuron-to-glia lipid transfer and astrocytic lipid droplet formation coupled with reactive oxygen species (ROS) accumulation. These changes caused astrocytes to uptake less glutamate thereby altering neuron excitability.
Conclusions: For a strong AD-associated locus near Clusterin (CLU), we connected an AD protective allele to a role of neuronal CLU in promoting neuron excitability through lipid-mediated neuron-glia communication. Our study provides insights into how CLU confers resilience to AD through neuron-glia interactions.
Keywords: Allele-specific open chromatin; Alzheimer’s disease; Clusterin; Genome-wide association study; IPSC; Lipid droplets; Neuron excitability; Neuron-glia lipid transfer; Protective allele.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Any data derived from human cells and tissue, and mouse experiments were approved by Endeavor Health (formerly NorthShore University HealthSystem) institutional review board (IRB). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Update of
-
Alzheimer's disease protective allele of Clusterin modulates neuronal excitability through lipid-droplet-mediated neuron-glia communication.medRxiv [Preprint]. 2024 Aug 15:2024.08.14.24312009. doi: 10.1101/2024.08.14.24312009. medRxiv. 2024. Update in: Mol Neurodegener. 2025 May 3;20(1):51. doi: 10.1186/s13024-025-00840-1. PMID: 39185522 Free PMC article. Updated. Preprint.
Similar articles
-
Alzheimer's disease protective allele of Clusterin modulates neuronal excitability through lipid-droplet-mediated neuron-glia communication.medRxiv [Preprint]. 2024 Aug 15:2024.08.14.24312009. doi: 10.1101/2024.08.14.24312009. medRxiv. 2024. Update in: Mol Neurodegener. 2025 May 3;20(1):51. doi: 10.1186/s13024-025-00840-1. PMID: 39185522 Free PMC article. Updated. Preprint.
-
Clusterin secreted from astrocyte promotes excitatory synaptic transmission and ameliorates Alzheimer's disease neuropathology.Mol Neurodegener. 2021 Jan 31;16(1):5. doi: 10.1186/s13024-021-00426-7. Mol Neurodegener. 2021. PMID: 33517893 Free PMC article.
-
Astrocytic response mediated by the CLU risk allele inhibits OPC proliferation and myelination in a human iPSC model.Cell Rep. 2023 Aug 29;42(8):112841. doi: 10.1016/j.celrep.2023.112841. Epub 2023 Jul 25. Cell Rep. 2023. PMID: 37494190 Free PMC article.
-
Exploring the Role of CLU in the Pathogenesis of Alzheimer's Disease.Neurotox Res. 2021 Dec;39(6):2108-2119. doi: 10.1007/s12640-020-00271-4. Epub 2020 Aug 21. Neurotox Res. 2021. Retraction in: Neurotox Res. 2022 Aug;40(4):1125. doi: 10.1007/s12640-022-00519-1. PMID: 32820456 Retracted. Review.
-
The role of clusterin in Alzheimer's disease: pathways, pathogenesis, and therapy.Mol Neurobiol. 2012 Apr;45(2):314-26. doi: 10.1007/s12035-012-8237-1. Epub 2012 Jan 25. Mol Neurobiol. 2012. PMID: 22274961 Review.
Cited by
-
Single-cell multiomics of neuronal activation reveals context-dependent genetic control of brain disorders.bioRxiv [Preprint]. 2025 Feb 17:2025.02.17.638682. doi: 10.1101/2025.02.17.638682. bioRxiv. 2025. PMID: 40027724 Free PMC article. Preprint.
-
CRISPR/Cas9 and iPSC-Based Therapeutic Approaches in Alzheimer's Disease.Antioxidants (Basel). 2025 Jun 25;14(7):781. doi: 10.3390/antiox14070781. Antioxidants (Basel). 2025. PMID: 40722885 Free PMC article. Review.
References
-
- Scheltens P, Blennow K, Breteler MM, de Strooper B, Frisoni GB, Salloway S, Van der Flier WM. Alzheimer’s disease. Lancet. 2016;388:505–17. - PubMed
MeSH terms
Substances
Grants and funding
- RF1AG079141/AG/NIA NIH HHS/United States
- U01 AG072439/AG/NIA NIH HHS/United States
- R01 MH116281/MH/NIMH NIH HHS/United States
- RF1 AG079141/AG/NIA NIH HHS/United States
- R01AG063175/AG/NIA NIH HHS/United States
- R01 AG063175/AG/NIA NIH HHS/United States
- R01MH106575/National Institute of Mental Health and Neurosciences
- R01 MH106575/MH/NIMH NIH HHS/United States
- R01 AG073260/AG/NIA NIH HHS/United States
- R01AG081374/AG/NIA NIH HHS/United States
- R01MH116281/National Institute of Mental Health and Neurosciences
- R01 AG081374/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

