DNA methylation in primary myelofibrosis is partly associated with driver mutations and distinct from other myeloid malignancies
- PMID: 40319310
- PMCID: PMC12048995
- DOI: 10.1186/s13148-025-01877-1
DNA methylation in primary myelofibrosis is partly associated with driver mutations and distinct from other myeloid malignancies
Abstract
Background: Primary myelofibrosis (PMF) is a clonal blood disorder characterized by mutually exclusive driver mutations in JAK2, CALR, or MPL genes. So far, it is largely unclear if the driver mutations have a specific impact on DNA methylation (DNAm) profiles and how epigenetic alterations in PMF are related to other myeloid malignancies.
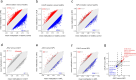
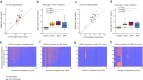
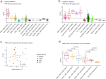
Results: When we compared DNAm profiles from PMF patients we found very similar epigenetic modifications in JAK2 and CALR mutated cases, whereas MPL mutations displayed less pronounced and distinct patterns. Furthermore, induced pluripotent stem cell (iPSC) models with JAK2 mutations indicated only a moderate association with PMF-related epigenetic changes, suggesting that these alterations may not be directly driven by the mutations themselves. Additionally, PMF-associated epigenetic changes showed minimal correlation with allele burden and seemed to be largely influenced by shifts in the cellular composition. PMF DNAm profiles compared with those from other myeloid malignancies-such as acute myeloid leukemia, juvenile myelomonocytic leukemia, and myelodysplastic syndrome-showed numerous overlapping changes, making it difficult to distinguish PMF based on individual CpGs. However, a PMF score created by combining five CpGs was able to discern PMF from other diseases.
Conclusion: These findings demonstrate that PMF driver mutations do not directly evoke epigenetic changes. While PMF shares epigenetic alterations with other myeloid malignancies, DNA methylation patterns can distinguish between PMF and related diseases.
Keywords: AML; CpG; DNA methylation; Epigenetic; JMML; MDS; MPN; Myeloid malignancies; Primary myelofibrosis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All samples were obtained after informed and written consent in accordance with the Declaration of Helsinki and the research was specifically approved by the local ethics committee of RWTH Aachen University (EK 041/15, EK 206/09 and EK 127/12). Consent for publication: Not applicable. Competing interests: W.W. and V.T. are involved in the company Cygenia GmbH ( www.cygenia.com ) that can provide service for epigenetic analysis to other scientists. Apart from this the authors have no competing interests to declare.
Figures




Similar articles
-
JAK2 V617F, MPL, and CALR Mutations in Korean Patients with Essential Thrombocythemia and Primary Myelofibrosis.J Korean Med Sci. 2015 Jul;30(7):882-8. doi: 10.3346/jkms.2015.30.7.882. Epub 2015 Jun 10. J Korean Med Sci. 2015. PMID: 26130950 Free PMC article.
-
CALR, JAK2, and MPL mutation profiles in patients with four different subtypes of myeloproliferative neoplasms: primary myelofibrosis, essential thrombocythemia, polycythemia vera, and myeloproliferative neoplasm, unclassifiable.Am J Clin Pathol. 2015 May;143(5):635-44. doi: 10.1309/AJCPUAAC16LIWZMM. Am J Clin Pathol. 2015. PMID: 25873496
-
Clinical effect of driver mutations of JAK2, CALR, or MPL in primary myelofibrosis.Blood. 2014 Aug 14;124(7):1062-9. doi: 10.1182/blood-2014-05-578435. Epub 2014 Jul 1. Blood. 2014. PMID: 24986690 Free PMC article.
-
Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms.Blood. 2017 Feb 9;129(6):667-679. doi: 10.1182/blood-2016-10-695940. Epub 2016 Dec 27. Blood. 2017. PMID: 28028029 Review.
-
Presence of triple positive driver mutations in JAK2, CALR and MPL in primary myelofibrosis: a case report and literature review.Hematology. 2024 Dec;29(1):2402106. doi: 10.1080/16078454.2024.2402106. Epub 2024 Sep 13. Hematology. 2024. PMID: 39268974 Review.
References
-
- Tefferi A, Lasho TL, Finke CM, Knudson RA, Ketterling R, Hanson CH, et al. CALR vs JAK2 vs MPL-mutated or triple-negative myelofibrosis: clinical, cytogenetic and molecular comparisons. Leukemia. 2014;28(7):1472–7. - PubMed
-
- Tefferi A, Pardanani A. Myeloproliferative neoplasms: a contemporary review. JAMA Oncol. 2015;1(1):97–105. - PubMed
-
- Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. New Engl J Med. 2013;369(25):2379–90. - PubMed
-
- Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31(5):274–80. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

