Identification and characterization of tertiary lymphoid structures in brain metastases
- PMID: 40319321
- PMCID: PMC12049775
- DOI: 10.1186/s40478-025-02007-x
Identification and characterization of tertiary lymphoid structures in brain metastases
Abstract
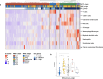
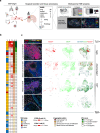
Brain metastases (BrM) are the most common cancers in the brain and linked to poor prognosis. Given the high incidence and often limited treatment options, understanding the complexity of the BrM tumor microenvironment is crucial for the development of novel therapeutic strategies. We performed transcriptome-wide gene expression profiling combined with spatial immune cell profiling to characterize the tumor immune microenvironment in 95 patients with BrM from different primary tumors. We found that BrM from lung carcinoma and malignant melanoma showed overall higher immune cell infiltration as compared to BrM from breast carcinoma. RNA sequencing-based immune cell deconvolution revealed gene expression signatures indicative of tertiary lymphoid structures (TLS) in subsets of BrM, mostly from lung cancer and melanoma. This finding was corroborated by multiplex immunofluorescence staining of immune cells in BrM tissue sections. Detection of TLS signatures was more common in treatment-naïve BrM and associated with prolonged survival after BrM diagnosis in lung cancer patients. Our findings highlight the cellular diversity of the tumor immune microenvironment in BrM of different cancer types and suggest a role of TLS formation for BrM patient outcome.
Keywords: Brain metastasis; Immune cell deconvolution; Multiplex immunofluorescence; Tertiary lymphoid structures; Tumor microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: The patients gave their written informed consent for the use of their tissue samples and associated clinical data for research purposes. The study was approved by the institutional review board of the Medical Faculty, Heinrich Heine University Düsseldorf (study number: 5717). Competing interest: SSM, YR, IH, LM, JF, SS, JM, KK, HW, GR, KHP, BB declare no conflict of interest. VM: research support from Novartis, Roche, Seagen, Genentech, Astra Zeneca and honoraria for lectures from Astra Zeneca, arsTempi, Daiichi-Sankyo, Eisai, Pfizer, MSD, Medac, Novartis, Roche, Seagen, Onkowissen, high5 Oncology, Lilly, Medscape, Gilead, Pierre Fabre, iMED Institute as well as consultancy honoraria from Roche, Pierre Fabre, PINK, ClinSol, Novartis, MSD, Daiichi-Sankyo, Eisai, Lilly, Seagen, Gilead, Stemline. AB: research support from Daiichi Sankyo, Roche and honoraria for lectures, consultation or advisory board participation from Roche Bristol-Meyers Squibb, Merck, Daiichi Sankyo, AstraZeneca, CeCaVa, Seagen, Alexion, Servier as well as travel support from Roche, Amgen and AbbVie. TF: received honoraria from Onkowissen, FOMF, Medconcept and travel support from Roche. DS: has offered consultation to Philogen, lnFlarX, Neracare, Merck Sharp & Dohme, Novartis, Bristol Myers Squibb, Pfizer, Pierre Fabre, Replimune, SunPharma, Daiichi Sanyo, Astra Zeneca, IQVIA, LabCorp, UltimoVacs, Seagen, Immunocore, Immatics, BioNTech, PamGene, BioAlta, Regeneron, Agenus, Erasca, Formycon, NoviGenix, CureVac, and Sanofi and has received research grants from Amgen, BMS, MSD, and Pfizer. Informed consent: Not applicable.
Figures


References
-
- Achrol AS, Rennert RC, Anders C, Soffietti R, Ahluwalia MS, Nayak L, Peters S, Arvold ND, Harsh GR, Steeg PS, Chang SD (2019) Brain metastases. Nat Rev Dis Primers 5:5. 10.1038/s41572-018-0055-y - PubMed
-
- Barnholtz-Sloan JS, Sloan AE, Davis FG, Vigneau FD, Lai P, Sawaya RE (2004) Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the metropolitan detroit cancer surveillance system. JCO 22:2865–2872. 10.1200/JCO.2004.12.149 - PubMed
-
- Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman C, Fridman WH, De Reyniès A (2016) Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 17:218. 10.1186/s13059-016-1070-5 - PMC - PubMed
-
- Boire A, Brastianos PK, Garzia L, Valiente M (2020) Brain metastasis. Nat Rev Cancer 20:4–11. 10.1038/s41568-019-0220-y - PubMed
-
- Brastianos PK, Kim AE, Giobbie-Hurder A, Lee EQ, Lin NU, Overmoyer B, Wen PY, Nayak L, Cohen JV, Dietrich J, Eichler A, Heist RS, Krop I, Lawrence D, Ligibel J, Tolaney S, Mayer E, Winer E, Bent B, De Sauvage MA, Ijad N, Larson JM, Marion B, Nason S, Murthy N, Ratcliff S, Summers EJ, Mahar M, Shih HA, Oh K, Cahill DP, Gerstner ER, Sullivan RJ (2023) Pembrolizumab in brain metastases of diverse histologies: phase 2 trial results. Nat Med 29:1728–1737. 10.1038/s41591-023-02392-7 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

