Expanding the Spectrum of Canine Diffuse Large B-Cell Lymphoma Genetic Aberrations Through Whole Genome Sequencing Analysis
- PMID: 40320296
- PMCID: PMC12378081
- DOI: 10.1111/vco.13059
Expanding the Spectrum of Canine Diffuse Large B-Cell Lymphoma Genetic Aberrations Through Whole Genome Sequencing Analysis
Abstract
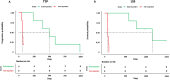
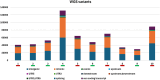
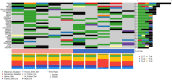
Diffuse large B-cell lymphoma (DLBCL) is one of the most prevalent haematological malignancies in both humans and dogs, characterised in both species by significant clinical heterogeneity and limited prognostic predictability. With the introduction of next-generation sequencing (NGS) technologies in veterinary medicine over the past decade, researchers have begun to elucidate the molecular basis of canine DLBCL (cDLBCL); however, much of the clinical heterogeneity associated with this tumour remains unexplained. In this study, we performed whole genome sequencing on 10 cDLBCL cases, all treated with chemo-immunotherapy, which exhibited similar clinico-pathological features but markedly different outcomes. Cases were classified as "poor" or "good" responders based on whether their lymphoma-specific survival fell below or above the cohort's median. Protein-coding variants and copy number aberrations unique to poor or good responders revealed novel candidate genes not previously identified in cDLBCL studies, while splicing, untranslated regions, and intronic variants were detected in genes already known to be recurrently mutated. In conclusion, our investigation has broadened the spectrum of potentially pathogenic variants implicated in cDLBCL, though further studies with larger cohorts are necessary to validate these findings.
Keywords: DLBCL; diffuse large B‐cell lymphoma; dog; prognosis; whole genome sequencing.
© 2025 The Author(s). Veterinary and Comparative Oncology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Characterization of the genomic landscape of canine diffuse large B-cell lymphoma reveals recurrent H3K27M mutations linked to progression-free survival.Sci Rep. 2025 Feb 8;15(1):4724. doi: 10.1038/s41598-025-89245-0. Sci Rep. 2025. PMID: 39922874 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Chimeric antigen receptor (CAR) T-cell therapy for people with relapsed or refractory diffuse large B-cell lymphoma.Cochrane Database Syst Rev. 2021 Sep 13;9(9):CD013365. doi: 10.1002/14651858.CD013365.pub2. Cochrane Database Syst Rev. 2021. PMID: 34515338 Free PMC article.
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

