This is a preprint.
Loss of CARM1 alters the developmental programming of Glioma stem-like cells and creates a druggable NGFR/NTRK dependency
- PMID: 40321217
- PMCID: PMC12047859
- DOI: 10.1101/2025.04.11.647869
Loss of CARM1 alters the developmental programming of Glioma stem-like cells and creates a druggable NGFR/NTRK dependency
Abstract
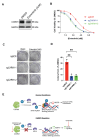
A key driver of Glioblastoma (GBM) heterogeneity and therapy resistance is the capacity of glioma stem-like cells (GSCs) to hijack developmental signaling programs. However, it remains unclear how GSCs regulate these adapted developmental signaling pathways and how these pathways might be therapeutically exploited. The arginine methyltransferase, CARM1, has been shown to play critical roles in maintaining stem cell pluripotency, preventing differentiation, and recently was discovered to be upregulated in Glioblastoma. To date, there is little to no understanding of the role that CARM1 plays in regulating developmental processes in Glioblastoma. To address this gap in knowledge, we applied a multi-omics approach to characterize developmental processes that are specifically regulated by CARM1 in GSCs. We found that loss of CARM1 results in dysregulation of several developmental markers: ARX, GFAP, NGFR, PDGFRA and results in both a proteomic and transcriptomic shift towards the radial glia cell lineage. Moreover, CARM1 depleted cells reprogram their signaling to develop an increased survival dependency on NGFR/NTRK signaling and are hypersensitive to the FDA approved brain penetrant NTRK inhibitor-Entrectinib. Mechanistically, we find that NFIA is a CARM1 substrate and can repress NGFR signaling just as CARM1 does, and thus the CARM1/NFIA relationship is likely a key regulator of NGFR/NTRK signaling in GSCs. Altogether, we demonstrate that CARM1 regulates the cell lineage of GSCs at the transcriptomic and proteomic level, and naturally represses NGFR/NTRK signaling-likely through CARM1 dependent methylation of NFIA. Further, CARM1 depletion leads GSCs to develop a survival dependency on NGFR/NTRK signaling and creates a therapeutic vulnerability to NTRK inhibition.
Keywords: AKT; CARM1; Capivasertib; Entrectinib; Glioblastoma; MAPK; NGFR; NTRK; arginine methylation; development; epigenetics; glioma stem-like cell; histones; proteomics; radial glial cell.
Conflict of interest statement
Conflict of interest statement The authors declare no financial conflicts of interest.
Figures





References
-
- Singh S.K., et al. , Identification of human brain tumour initiating cells. Nature, 2004. 432(7015): p. 396–401. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
