This is a preprint.
A genome-wide CRISPR screen identifies GRA38 as a key regulator of lipid homeostasis during Toxoplasma gondii adaptation to lipid-rich conditions
- PMID: 40321756
- PMCID: PMC12047978
- DOI: 10.21203/rs.3.rs-6436164/v1
A genome-wide CRISPR screen identifies GRA38 as a key regulator of lipid homeostasis during Toxoplasma gondii adaptation to lipid-rich conditions
Abstract
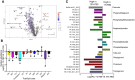
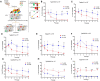
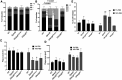
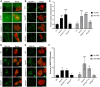
Intracellular parasites like Toxoplasma gondii scavenge host nutrients, particularly lipids, to support their growth and survival. Although Toxoplasma is known to adjust its metabolism based on nutrient availability, the mechanisms that mediate lipid sensing and metabolic adaptation remain poorly understood. Here, we performed a genome-wide CRISPR screen under lipid-rich (10% Fetal Bovine Serum (FBS)) and lipid-limited (1% FBS) conditions to identify genes critical for lipid-responsive fitness. We identified the Toxoplasma protein GRA38 as a lipid-dependent regulator of parasite fitness. GRA38 exhibits phosphatidic acid (PA) phosphatase (PAP) activity in vitro, which is significantly reduced by mutation of its conserved DxDxT/V catalytic motif. Disruption of GRA38 led to the accumulation of PA species and widespread alterations in lipid composition, consistent with impaired PAP activity. These lipid imbalances correlated with reduced parasite virulence in mice. Our findings identify GRA38 as a metabolic regulator important for maintaining lipid homeostasis and pathogenesis in Toxoplasma gondii.
Keywords: CRISPR screen; GRA38; Toxoplasma gondii; host-parasite metabolic interactions; lipidomics; metabolic adaptation; phosphatidic acid phosphatase.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest.
Figures

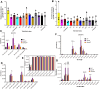

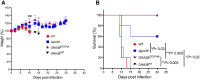
References
-
- Muñoz-Roldan M, Heimesaat MM, Liesenfeld O. Toxoplasmosis. In: Manson’s Tropical Infectious Diseases. Elsevier; 2014. p. 652–63.e1.
-
- Coppens I. Targeting lipid biosynthesis and salvage in apicomplexan parasites for improved chemotherapies. Nat Rev Microbiol. 2013. Dec;11(12):823–35. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

