Integrative bioinformatics analysis and experimental validation of key biomarkers driving the progression of cirrhotic portal hypertension
- PMID: 40321824
- PMCID: PMC12049105
- DOI: 10.7717/peerj.19360
Integrative bioinformatics analysis and experimental validation of key biomarkers driving the progression of cirrhotic portal hypertension
Abstract
Background: Portal hypertension is a driving factor of cirrhosis complications, but the specific molecular mechanism of portal hypertension in cirrhosis remains unclear. The aim of this study was to identify hub genes for predicting persistent progression of portal hypertension in patients with liver cirrhosis.
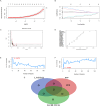
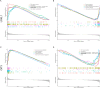
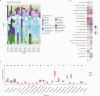
Methods: Related microarray datasets were obtained from the Gene Expression Omnibus database. Weighted gene co-expression network analysis and differential expression genes analysis were used to identify the correlation sets of genes. In addition, protein-protein interaction networks and machine learning algorithms were conducted to screen center of candidate genes. To validate the diagnostic effect of hub genes, receiver operating characteristic curves were utilized in another dataset that is publicly accessible. Furthermore, the CIBERSORT algorithm was employed to investigate the immune infiltration levels of 22 immune cells and their connection to hub gene markers. Immunohistochemistry and reverse transcription quantitative polymerase chain reaction (RT-qPCR) were conducted to validate novel hub genes in clinical specimens.
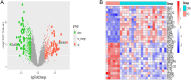
Results: We obtained 671 differentially expressed genes and 11 module genes related to cirrhotic portal hypertension. Two candidate genes namely oncoprotein-induced transcript 3 protein (OIT3) and lysyl oxidase like protein 1 (LOXL1) were identified as biomarkers. RT-qPCR and immunohistochemistry (IHC) verified the expression of LOXL1 and OIT3 at mRNA and protein levels in liver tissue.
Conclusions: OIT3 and LOXL1 were identified as potential novel targets for the diagnosis and treatment of cirrhotic portal hypertension (CPH).
Keywords: Bioinformatic analysis; Cirrhosis; Lysyl Oxidase Like Protein 1; Machine learning; Oncoprotein-induced Transcript 3 Protein; Portal hypertension; Weighted gene co-expression network analysis.
© 2025 Li et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Identifying hub genes of sepsis-associated and hepatic encephalopathies based on bioinformatic analysis-focus on the two common encephalopathies of septic cirrhotic patients in ICU.BMC Med Genomics. 2024 Jan 11;17(1):19. doi: 10.1186/s12920-023-01774-7. BMC Med Genomics. 2024. PMID: 38212812 Free PMC article.
-
Integrated bioinformatics combined with machine learning to analyze shared biomarkers and pathways in psoriasis and cervical squamous cell carcinoma.Front Immunol. 2024 May 28;15:1351908. doi: 10.3389/fimmu.2024.1351908. eCollection 2024. Front Immunol. 2024. PMID: 38863714 Free PMC article.
-
Identification of potential biomarkers of vascular calcification using bioinformatics analysis and validation in vivo.PeerJ. 2022 Mar 16;10:e13138. doi: 10.7717/peerj.13138. eCollection 2022. PeerJ. 2022. PMID: 35313524 Free PMC article.
-
Exploring the Immune Landscape of Cirrhosis through Weighted Gene Co-expression Network Analysis.Cell Mol Biol (Noisy-le-grand). 2023 Jun 30;69(6):168-174. doi: 10.14715/cmb/2023.69.6.25. Cell Mol Biol (Noisy-le-grand). 2023. PMID: 37605574
-
Identification of diagnostic genes and drug prediction in metabolic syndrome-associated rheumatoid arthritis by integrated bioinformatics analysis, machine learning, and molecular docking.Front Immunol. 2024 Jul 29;15:1431452. doi: 10.3389/fimmu.2024.1431452. eCollection 2024. Front Immunol. 2024. PMID: 39139563 Free PMC article.
References
-
- Csiszar K. Progress in Nucleic Acid Research and Molecular Biology. Cambridge, Massachusetts: Academic Press; 2001. Lysyl oxidases: a novel multifunctional amine oxidase family; pp. 1–32. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

