Identification of NUV-244 as a PNPLA3 I148M degrading small molecule
- PMID: 40322074
- PMCID: PMC12049818
- DOI: 10.1016/j.isci.2025.112384
Identification of NUV-244 as a PNPLA3 I148M degrading small molecule
Abstract
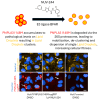
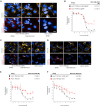
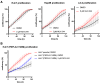
The PNPLA3 I148M variant is a key genetic determinant of metabolic dysfunction-associated steatotic liver disease (MASLD) and related conditions, contributing to lipid metabolism dysregulation and disease progression. To identify small molecules that modulate PNPLA3 I148M, we conducted a high-content screen of over 820,000 compounds and identified NUV-244, a potent degrader of PNPLA3 I148M in liver-derived cells. NUV-244 reduces PNPLA3 I148M levels on lipid droplets via the ubiquitin-proteasome system, involving the E3 ligase BFAR, without affecting PNPLA2. It restores lipid droplet morphology and improves cellular fitness in PNPLA3 I148M-expressing cells. These findings provide a tool to investigate PNPLA3 I148M function and offer a potential strategy for developing targeted therapies for MASLD and related diseases. By enabling selective degradation of PNPLA3 I148M, this approach expands therapeutic possibilities beyond genetic manipulation, addressing a critical need in metabolic liver disease research.
Keywords: Biochemistry; Biological sciences; Cell biology; Functional aspects of cell biology.
© 2025 The Authors.
Conflict of interest statement
P.S., N.B., V.P., N.Z., K.J., F.V.N., R.L., N.D., D.S., Z.M., F.K., S.S., B.B., and H.S. are or were employees of Nuvisan ICB GmbH, Berlin, Germany. M.H.B., J.S.P., L.B., and F.E.D. are or were employees of Foresite Labs, USA. S.F. and S.R. were consultants for Foresite Labs, USA.
Figures




References
-
- Cherubini A., Ostadreza M., Jamialahmadi O., Pelusi S., Rrapaj E., Casirati E., Passignani G., Norouziesfahani M., Sinopoli E., Baselli G., et al. Interaction between estrogen receptor-α and PNPLA3 p.I148M variant drives fatty liver disease susceptibility in women. Nat. Med. 2023;29:2643–2655. - PMC - PubMed
-
- Romeo S., Sanyal A., Valenti L. Leveraging Human Genetics to Identify Potential New Treatments for Fatty Liver Disease. Cell Metab. 2020;31:35–45. - PubMed
LinkOut - more resources
Full Text Sources

