Pathogen surveillance and risk factors for pulmonary infection in patients with lung cancer: A retrospective single-center study
- PMID: 40322468
- PMCID: PMC12048904
- DOI: 10.1515/med-2025-1180
Pathogen surveillance and risk factors for pulmonary infection in patients with lung cancer: A retrospective single-center study
Abstract
Background: Early and accurate diagnosis of pulmonary infection (PI) is crucial for the timely implementation of appropriate treatment strategies in lung cancer patients.
Methods: Metagenomic next-generation sequencing and conventional testing were performed in lung cancer patients with and without PI. The pathogen profiles were analyzed, and risk factors for PI were explored using univariate and multivariate logistic regression models.
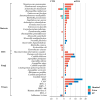
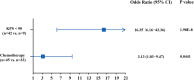
Results: A total of 55 lung cancer patients with PI and 59 non-infected lung cancer patients were included. There were 41 underlying pathogens identified by both methods in lung cancer patients with PI. The coexistence of different pathogen types was common, particularly between fungi and viruses, which was observed in 28.57% of cases. The incidence of Streptococcus pneumoniae and Pneumocystis jirovecii is significantly higher in small-cell lung carcinoma patients compared to that in non-small-cell lung carcinoma patients. Besides, cytomegalovirus, P. jirovecii, and Aspergillus were more likely to be found in advanced-stage patients. Risk factor analysis revealed that Karnofsky Performance Status <90 and chemotherapy were strongly associated with PI in lung cancer patients.
Conclusions: This study highlights the complexity of PI in lung cancer patients, emphasizing the need for tailored diagnostic and therapeutic strategies based on cancer type and stage.
Keywords: co-infection; lung cancer; mNGS; pathogens; pulmonary infection.
© 2025 the author(s), published by De Gruyter.
Conflict of interest statement
Conflict of interest: The authors state no conflict of interest.
Figures


Similar articles
-
Metagenomic next-generation sequencing targeted and metagenomic next-generation sequencing for pulmonary infection in HIV-infected and non-HIV-infected individuals.Front Cell Infect Microbiol. 2024 Aug 19;14:1438982. doi: 10.3389/fcimb.2024.1438982. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39224706 Free PMC article.
-
A single-center, retrospective study of hospitalized patients with lower respiratory tract infections: clinical assessment of metagenomic next-generation sequencing and identification of risk factors in patients.Respir Res. 2024 Jun 20;25(1):250. doi: 10.1186/s12931-024-02887-y. Respir Res. 2024. PMID: 38902783 Free PMC article.
-
Microbiomes Detected by Bronchoalveolar Lavage Fluid Metagenomic Next-Generation Sequencing among HIV-Infected and Uninfected Patients with Pulmonary Infection.Microbiol Spectr. 2023 Aug 17;11(4):e0000523. doi: 10.1128/spectrum.00005-23. Epub 2023 Jul 12. Microbiol Spectr. 2023. PMID: 37436163 Free PMC article.
-
Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis.BMC Pulm Med. 2019 Dec 19;19(1):252. doi: 10.1186/s12890-019-1022-4. BMC Pulm Med. 2019. PMID: 31856779 Free PMC article.
-
Diagnostic value of metagenomic next-generation sequencing of bronchoalveolar lavage fluid for the diagnosis of suspected pneumonia in immunocompromised patients.BMC Infect Dis. 2022 Apr 29;22(1):416. doi: 10.1186/s12879-022-07381-8. BMC Infect Dis. 2022. PMID: 35488253 Free PMC article.
References
-
- Chhikara BS, Parang K. Global Cancer Statistics 2022: the trends projection analysis. Chem Biol Lett. 2023;10(1):451.
-
- Sarihan S, Ercan I, Saran A, Cetintas SK, Akalin H, Engin K. Evaluation of infections in non-small cell lung cancer patients treated with radiotherapy. Cancer Detect Prev. 2005;29(2):181–8. - PubMed
-
- Vento S, Cainelli F, Temesgen Z. Lung infections after cancer chemotherapy. Lancet Oncol. 2008;9(10):982–92. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
