Diving into Diversity: Hasleaberepwari (Bacillariophyceae, Naviculaceae), a new species of marine diatom from New Caledonia
- PMID: 40322669
- PMCID: PMC12048817
- DOI: 10.3897/phytokeys.255.144697
Diving into Diversity: Hasleaberepwari (Bacillariophyceae, Naviculaceae), a new species of marine diatom from New Caledonia
Abstract
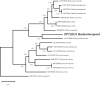
The current article introduces and describes Hasleaberepwari sp. nov., a new species of diatom discovered in the vicinity of Boulouparis, New Caledonia. Under light microscopy, H.berepwari sp. nov. strongly resembles Hasleapseudostrearia, but preliminary molecular barcoding conducted using partial 18S and rbcL genes suggested that it was a distinct species. This was confirmed first by scanning electron microscopy which showed the differences in stria densities between both species. A short-reads genome-skimming protocol applied on H.berepwari sp. nov. led us to obtain its complete mitochondrial and plastid genomes. The mitogenome is 36,572 bp in length and as already observed among other species of Haslea spp., the nad6 and nad2 genes are fused within a single open-reading frame. The plastome is 131,897 bp length, and unlike the mitogenome, it is not colinear with those of H.pseudostrearia. The results derived from the sequencing of the plastome allowed to perform a 123-gene multigene maximum likelihood phylogeny that associates H.berepwari sp. nov. to H.pseudostrearia with maximum support at the nodes but also strictly distinguishes them, suggesting a greater genetic distance between these species than what has been previously observed between other marennine-producing species.
Keywords: Coral Sea; Naviculales; organellar genomes; tropical diatoms.
Fiddy Semba Prasetiya, Thierry Jauffrais, Mustapha Mohamed-Benkada, Nolwenn Callac, Dominique Ansquer, Elif Yılmaz, Claude Lemieux, Monique Turmel, Jean-Luc Mouget, Danang Ambar Prabowo, Debora Christin Purbani, Diah Radini Noerdjito, Varian Fahmi, Sulastri Arsad, Romain Gastineau.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Adjout R, Mouget JL, Pruvost J, Chentir I, Loiseau C, Baba Hamed MB. (2022) Effects of temperature, irradiance, and pH on the growth and biochemical composition of Hasleaostrearia batch-cultured in an airlift plan-photobioreactor. Applied Microbiology and Biotechnology 106(13–16): 5233–5247. 10.1007/s00253-022-12055-1 - DOI - PubMed
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov A, Lesin VM, Nikolenko SI, Pham S, Prjibelski A, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. (2012) SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19: 455–477. 10.1089/cmb.2012.0021 - DOI - PMC - PubMed
-
- Bell JD, Agudo NN, Purcell SW, Blazer P, Simutoga M, Pham D, Della Patrona L. (2007) Grow-out of sandfish Holothuriascabra in ponds shows that co-culture with shrimp Litopenaeusstylirostris is not viable. Aquaculture (Amsterdam, Netherlands) 273(4): 509–519. 10.1016/j.aquaculture.2007.07.015 - DOI
-
- Callac N, Boulo V, Giraud C, Beauvais M, Ansquer D, Ballan V, Maillez JR, Wabete N, Pham D. (2022) Microbiota of the rearing water of Penaeusstylirostris larvae influenced by lagoon seawater and specific key microbial lineages of larval stage and survival. Microbiology Spectrum 10(6): e0424122. 10.1128/spectrum.04241-22 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
