Melanoma Proteomics Unveiled: Harmonizing Diverse Data Sets for Biomarker Discovery and Clinical Insights via MEL-PLOT
- PMID: 40322912
- PMCID: PMC12150306
- DOI: 10.1021/acs.jproteome.4c00749
Melanoma Proteomics Unveiled: Harmonizing Diverse Data Sets for Biomarker Discovery and Clinical Insights via MEL-PLOT
Abstract
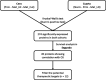
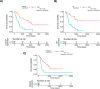
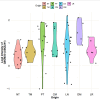
Using several melanoma proteomics data sets we created a single analysis platform that enables the discovery, knowledge build, and validation of diagnostic, predictive, and prognostic biomarkers at the protein level. Quantitative mass-spectrometry-based proteomic data was obtained from five independent cohorts, including 489 tissue samples from 394 patients with accompanying clinical metadata. We established an interactive R-based web platform that enables the comparison of protein levels across diverse cohorts, and supports correlation analysis between proteins and clinical metadata including survival outcomes. By comparing differential protein levels between metastatic, primary tumor, and nonmalignant samples in two of the cohorts, we identified 274 proteins showing significant differences among the sample types. Further analysis of these 274 proteins in lymph node metastatic samples from a third cohort revealed that 45 proteins exhibited a significant effect on patient survival. The three most significant proteins were HP (HR = 4.67, p = 2.8e-06), LGALS7 (HR = 3.83, p = 2.9e-05), and UBQLN1 (HR = 3.2, p = 4.8e-05). The user-friendly interactive web platform, accessible at https://www.tnmplot.com/melanoma, provides an interactive interface for the analysis of proteomic and clinical data. The MEL-PLOT platform, through its interactive capabilities, streamlines the creation of a comprehensive knowledge base, empowering hypothesis formulation and diligent monitoring of the most recent advancements in the domains of biomedical research and drug development.
Keywords: mass spectrometry; proteomics; skin cancer; survival; tumor progression.
Figures

References
-
- Surman M., Kedracka-Krok S., Hoja-Lukowicz D., Jankowska U., Drozdz A., Stepien E. L., Przybylo M.. Mass Spectrometry-Based Proteomic Characterization of Cutaneous Melanoma Ectosomes Reveals the Presence of Cancer-Related Molecules. International Journal of Molecular Sciences. 2020;21(8):2934. doi: 10.3390/ijms21082934. - DOI - PMC - PubMed
-
- Fekete J. T., Gyorffy B.. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3,104 breast cancer patients. Int. J. Cancer. 2019;145(11):3140–3151. doi: 10.1002/ijc.32369. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

