WashU Epigenome Browser update 2025
- PMID: 40322916
- PMCID: PMC12230663
- DOI: 10.1093/nar/gkaf387
WashU Epigenome Browser update 2025
Abstract
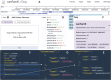
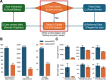
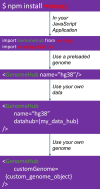
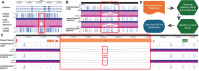
The WashU Epigenome Browser (https://epigenomegateway.wustl.edu/) is a web-based tool for exploring genomic data and providing visualization, investigation, and analysis of epigenomic datasets. Since its 2018 update, the redesigned user interface and newly developed features have enhanced how investigators interact with both the Browser and the extensive genomic data it hosts. The rapid evolution of the JavaScript ecosystem has presented new challenges and opportunities in maintaining and developing the WashU Epigenome Browser. In this update, we present a completely rewritten codebase. This new codebase minimizes the use of external libraries whenever possible, resulting in a significantly smaller code bundle size after production compilation. The reduced code size improves loading efficiency and boosts the Browser's performance, with improved scripting, graphics rendering, and painting performance. Lowering external dependencies also allows for faster and more straightforward installation. Additionally, the update includes a redesign of the user interface to further enhance user experience and features a new modular design in the codebase that enables the Browser to be exported as stand-alone modules for use in other web applications. Several novel track types for long-read methylation data and single-cell methylation data visualization have been added, and we continue to update and expand the data hubs we host for major consortia. We constructed the first data hub to systematically compare genomic data mapped to different genome assemblies, focusing on comparisons between hg38 and the first human T2T genome, chm13, using our new comparative genomics track function. The WashU Epigenome Browser also serves as a foundation for other genomics platforms, such as the WashU Virus Genome Browser, developed for SARS-COV-2 research, the WashU Comparative Epigenome Browser, and the WashU Repeat Browser.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures


References
-
- Lander ES, Linton LM, Birren B et al. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. - PubMed
MeSH terms
Grants and funding
- U01 HG009391/HG/NHGRI NIH HHS/United States
- UM1HG011585/HG/NHGRI NIH HHS/United States
- U01HG009391/HG/NHGRI NIH HHS/United States
- U41 HG010972/HG/NHGRI NIH HHS/United States
- UM1MH130994/HG/NHGRI NIH HHS/United States
- U01 CA200060/CA/NCI NIH HHS/United States
- U24HG012070/HG/NHGRI NIH HHS/United States
- UM1 HG011585/HG/NHGRI NIH HHS/United States
- U01CA200060/HG/NHGRI NIH HHS/United States
- U41HG010972/HG/NHGRI NIH HHS/United States
- UM1 MH130994/MH/NIMH NIH HHS/United States
- U24 HG012070/HG/NHGRI NIH HHS/United States
- R01 HG007175/HG/NHGRI NIH HHS/United States
- R01HG007175/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

