Epigenetic background of lineage-specific gene expression landscapes of four Staphylococcus aureus hospital isolates
- PMID: 40323905
- PMCID: PMC12052166
- DOI: 10.1371/journal.pone.0322006
Epigenetic background of lineage-specific gene expression landscapes of four Staphylococcus aureus hospital isolates
Abstract
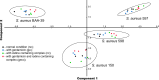
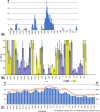
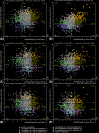
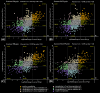
Bacteria with similar genomes can exhibit different phenotypes due to alternative gene expression patterns. In this study, we analysed four antibiotic-resistant Staphylococcus aureus hospital isolates using transcriptomics, PacBio genome sequencing, and methylomics analyses. Transcriptomic data were obtained from cultures exposed to gentamicin, the iodine-alanine complex CC-196, and their combination. We observed strain-specific expression patterns of core and accessory genes that remained stable under antimicrobial stress - a phenomenon we term the Clonal Gene Expression Stability (CGES) that is the main discovery of the paper. An involvement of epigenetic mechanisms in stabilization of the CGES was hypothesized and statistically verified. Canonical methylation patterns controlled by type I restriction-modification systems accounted for ~ 10% of epigenetically modified adenine residues, whereas multiple non-canonically modified adenines were distributed sporadically due to imperfect DNA targeting by methyltransferases. Protein-coding sequences were characterized by a significantly lower frequency of modified nucleotides. Epigenetic modifications near transcription start codons showed a statistically significant negative association with gene expression levels. While the role of epigenetic modifications in gene regulation remains debatable, variations in non-canonical modification patterns may serve as markers of CGES.
Copyright: © 2025 Korotetskiy et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Dominelli N, Jäger HY, Langer A, Brachmann A, Heermann R. High-throughput sequencing analysis reveals genomic similarity in phenotypic heterogeneous Photorhabdus luminescens cell populations. Ann Microbiol. 2022;72(1):20. doi: 10.1186/s13213-022-01677-5 - DOI
-
- Sánchez-Romero MA, Casadesús J. The bacterial epigenome. Nat Rev Microbiol. 2020;18(1):7–20. doi: 10.1038/s41579-019-0286-2. - PubMed
-
- Vasilchenko NG, Prazdnova EV, Lewitin E. Epigenetic mechanisms of gene expression regulation in bacteria of the Genus Bacillus. Russian J Gen. 2022;58:(1):1–19. doi: 10.1134/s1022795422010124 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

