The non-specific phospholipase C of common bean PvNPC4 modulates roots and nodule development
- PMID: 40323933
- PMCID: PMC12052164
- DOI: 10.1371/journal.pone.0306505
The non-specific phospholipase C of common bean PvNPC4 modulates roots and nodule development
Abstract
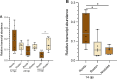
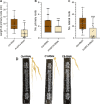
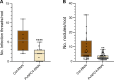
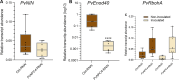
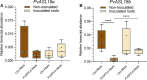
Plant phospholipase C (PLC) proteins are phospholipid-degrading enzymes classified into two subfamilies: phosphoinositide-specific PLCs (PI-PLCs) and non-specific PLCs (NPCs). PI-PLCs have been widely studied in various biological contexts, including responses to abiotic and biotic stresses and plant development; NPCs have been less thoroughly studied. No PLC subfamily has been characterized in relation to the symbiotic interaction between Fabaceae (legume) species and the nitrogen-fixing bacteria called rhizobia. However, lipids are reported to be crucial to this interaction, and PLCs may therefore contribute to regulating legume-rhizobia symbiosis. In this work, we functionally characterized NPC4 from common bean (Phaseolus vulgaris L.) during rhizobial symbiosis, findings evidence that NPC4 plays an important role in bean root development. The knockdown of PvNPC4 by RNA interference (RNAi) resulted in fewer and shorter primary roots and fewer lateral roots than were seen in control plants. Importantly, this phenotype seems to be related to altered auxin signaling. In the bean-rhizobia symbiosis, PvNPC4 transcript abundance increased 3 days after inoculation with Rhizobium tropici. Moreover, the number of infection threads and nodules, as well as the transcript abundance of PvEnod40, a regulatory gene of early stages of symbiosis, decreased in PvNPC4-RNAi roots. Additionally, transcript abundance of genes involved in autoregulation of nodulation (AON) was altered by PvNPC4 silencing. These results indicate that PvNPC4 is a key regulator of root and nodule development, underscoring the participation of PLC in rhizobial symbiosis.
Copyright: © 2025 Pacheco et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

