Real-World Genomic Landscape of Gastrointestinal Cancers in Asia and the Middle East Using Comprehensive Circulating Tumor DNA Next-Generation Sequencing
- PMID: 40324346
- PMCID: PMC12316443
- DOI: 10.1159/000545560
Real-World Genomic Landscape of Gastrointestinal Cancers in Asia and the Middle East Using Comprehensive Circulating Tumor DNA Next-Generation Sequencing
Abstract
Introduction: Gastrointestinal malignancies account for 25% of all cancer cases and 35% of cancer-related mortality. Next-generation sequencing (NGS) can elucidate the genomic landscape of gastrointestinal cancers; tissue-based genotyping has traditionally been used, but liquid biopsy-based genotyping is a noninvasive alternative. Moreover, geographical variations in the genomic landscape of gastrointestinal cancers have not been fully elucidated. This retrospective study aimed to gain insight into the genomic landscape of patients with gastrointestinal cancers from the Asia and Middle East (AME) region using plasma-derived circulating tumor DNA (ctDNA).
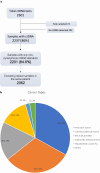
Methods: From routine clinical practice, 2,601 plasma samples were collected from 2,062 patients with gastrointestinal cancers in the AME region. NGS profiling was conducted using the Guardant360® assay. The frequency of biomarkers that can aid decision-making in cancer patients was investigated.
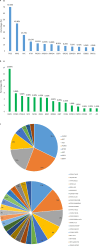
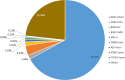
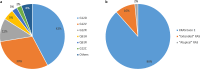
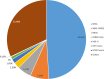
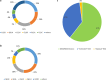
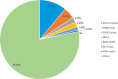
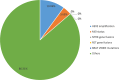
Results: Single-nucleotide variants affected most commonly TP53 (70.4%), KRAS (44.0%), APC (25.7%), ATM (15.1%), and PIK3CA (12.3%). Copy number alterations were most often observed in EGFR (13.7%), CCNE1 (5.9%), PIK3CA (5.0%), MYC (4.7%), and FGFR1 (4.6%); fusions were detected in 1.6% of patients and most frequently affected FGFR2, RET, ALK, FGFR3, and NTRK1/3. In patients with pancreatic adenocarcinoma, the most frequently observed clinically informative genomic biomarkers occurred in KRAS (G12C, 1.6%; all others, 67.1%), BRCA1/2 (4.1%), BRAF (V600X, 1.5%), and microsatellite instability-high (MSI-H) (1.0%). In patients with colorectal cancer, the most common clinically relevant alterations were KRAS (49.0%), BRAF (V600E, 7.6%), and NRAS (5.7%) mutations; ERBB2 amplifications (2.5%); and MSI-H (1.8%). In patients with biliary tract cancers, actionable alterations included IDH1 mutations (11.1%), ERBB2 amplifications (4.6%), FGFR2 fusions (2.0%), MSI-H (2.0%), and BRAF V600E (1.5%). In patients with gastric or gastroesophageal junction adenocarcinomas, actionable alterations included ERBB2 amplifications (10.1%) and MSI-H (3.6%).
Conclusion: Our data provide insight into the genomic landscape of patients with gastrointestinal cancers from the AME region using ctDNA analysis. These findings highlight the potential utility of liquid biopsy as a noninvasive tool for characterizing tumor genomic profiles and support its role in clinical practice.
Keywords: Circulating tumor DNA; Gastrointestinal cancers; Genomic landscape; Liquid biopsy.
© 2025 The Author(s). Published by S. Karger AG, Basel.
Conflict of interest statement
M.M. declares collaborations with Chugai, Taiho, NTT, PRiME-R, Canon Medical, Intage Healthcare, HU holdings, NTT data, XCOO, and Meiji Seika Pharma and being an invited speaker for Novartis, Daiichi Sankyo, MSD, Shionogi, Yakult, Merck, and Takeda. Y.S. declares research funding from Chugai Pharmaceutical Co., Ltd. and Taiho Pharmaceutical Co., Ltd.; honoraria from Eli Lilly Japan K.K., Bristol-Myers Squibb K.K., Chugai Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ono Pharmaceutical Co., Ltd., Merck Biopharma Co., Ltd., Taiho Pharmaceutical Co., Ltd., Bayer Yakuhin, Ltd., Daiichi Sankyo Co., Ltd., MSD K.K., Novartis Pharmaceuticals, Astellas Pharma Inc., Sysmex, and Guardant Health; and participation on a Data Safety Monitoring Board or Advisory Board for Merck Biopharma Co., Ltd., Ono Pharmaceutical Co., Ltd., and Guardant Health. N.S. and S.O. declare employment at Guardant Health AMEA, Inc. and stock ownership of Guardant Health, Inc. S.D. declares honoraria from Novartis, Pfizer, Lilly, AstraZeneca, Roche, MSD, BMS, New Bridge, and Caris as well as research funding from MSD, Amgen, and the Al Jalila Foundation. N.R. declares employment from Fortis Memorial Research Institute; stock and other ownership interests from Datar Cancer Genetics; honoraria from Lilly, Roche India, and Guardant Health AMEA; a consulting or advisory role at Guardant Health AMEA; and travel, accommodations, expenses from Guardant Health AMEA. The other authors have no conflicts of interest to declare.
Figures
Similar articles
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Characterization of the Thyroid Cancer Genomic Landscape by Plasma-Based Circulating Tumor DNA Next-Generation Sequencing.Thyroid. 2024 Feb;34(2):197-205. doi: 10.1089/thy.2023.0204. Epub 2023 Dec 22. Thyroid. 2024. PMID: 37962267
-
Elucidating the Role of KRAS, NRAS, and BRAF Mutations and Microsatellite Instability in Colorectal Cancer via Next-Generation Sequencing.Cancers (Basel). 2025 Jun 20;17(13):2071. doi: 10.3390/cancers17132071. Cancers (Basel). 2025. PMID: 40647370 Free PMC article.
-
Molecular testing for Lynch syndrome in people with colorectal cancer: systematic reviews and economic evaluation.Health Technol Assess. 2017 Sep;21(51):1-238. doi: 10.3310/hta21510. Health Technol Assess. 2017. PMID: 28895526 Free PMC article.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

