Cytochrome c oxidase inactivation in Physcomitrium patens reveals that respiration coordinates plant metabolism
- PMID: 40324757
- PMCID: PMC12164586
- DOI: 10.1093/plcell/koaf101
Cytochrome c oxidase inactivation in Physcomitrium patens reveals that respiration coordinates plant metabolism
Abstract
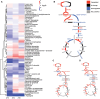
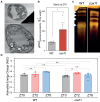
Photosynthetic organisms use sunlight as an energy source but rely on respiration during the night and in nonphotosynthetic tissues. Respiration also occurs in photosynthetically active cells, where its role is still unclear due to the lack of viable mutants. Mutations abolishing cytochrome c oxidase (Complex IV) activity are generally lethal. In this study, we generated cytochrome c oxidase assembly protein 11 (cox11) knockout lines through vegetative propagation in the moss Physcomitrium patens. These mutants showed severely impaired growth, with an altered composition of the respiratory apparatus and increased electron transfer through alternative oxidase. The light phase of photosynthesis remained largely unaffected in cox11 plants, while the efficiency of carbon fixation was reduced. Transcriptomic and metabolomic analyses showed that disrupting the cytochrome pathway had broad consequences for carbon and nitrogen metabolism. A major alteration in nitrogen assimilation was observed, with a general reduction in amino acid abundance. Partial growth rescue was achieved by externally supplying plants with amino acids but not with sugars, demonstrating that respiration in photosynthetic plant cells plays an essential role at the interface between carbon and nitrogen metabolism and a key role in providing carbon skeletons for amino acid biosynthesis.
© The Author(s) 2025. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

