A key residue of the extracellular gate provides quality control contributing to ABCG substrate specificity
- PMID: 40324983
- PMCID: PMC12052975
- DOI: 10.1038/s41467-025-59518-3
A key residue of the extracellular gate provides quality control contributing to ABCG substrate specificity
Abstract
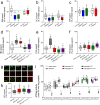
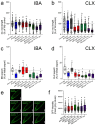
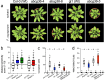
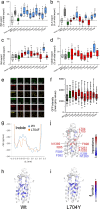
For G-type ATP-binding cassette (ABC) transporters, a hydrophobic "di-leucine motif" as part of a hydrophobic extracellular gate has been described to separate a large substrate-binding cavity from a smaller upper cavity and proposed to act as a valve controlling drug extrusion. Here, we show that an L704F mutation in the hydrophobic extracellular gate of Arabidopsis ABCG36/PDR8/PEN3 uncouples the export of the auxin precursor indole-3-butyric acid (IBA) from that of the defense compound camalexin (CLX). Molecular dynamics simulations reveal increased free energy for CLX translocation in ABCG36L704F and reduced CLX contacts within the binding pocket proximal to the extracellular gate region. Mutation L704Y enables export of structurally related non-ABCG36 substrates, IAA, and indole, indicating allosteric communication between the extracellular gate and distant transport pathway regions. An evolutionary analysis identifies L704 as a Brassicaceae family-specific key residue of the extracellular gate that controls the identity of chemically similar substrates. In summary, our work supports the conclusion that L704 is a key residue of the extracellular gate that provides a final quality control contributing to ABCG substrate specificity, allowing for balance of growth-defense trade-offs.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Ambudkar, S. V., Kimchi-Sarfaty, C., Sauna, Z. E. & Gottesman, M. M. P-glycoprotein: from genomics to mechanism. Oncogene22, 7468–7485 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

