A machine learning approach for multimodal data fusion for survival prediction in cancer patients
- PMID: 40325104
- PMCID: PMC12053085
- DOI: 10.1038/s41698-025-00917-6
A machine learning approach for multimodal data fusion for survival prediction in cancer patients
Abstract
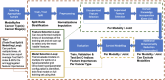
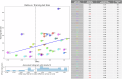
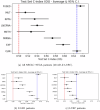
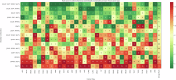
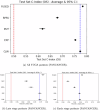
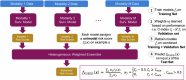
Technological advancements of the past decade have transformed cancer research, improving patient survival predictions through genotyping and multimodal data analysis. However, there is no comprehensive machine-learning pipeline for comparing methods to enhance these predictions. To address this, a versatile pipeline using The Cancer Genome Atlas (TCGA) data was developed, incorporating various data modalities such as transcripts, proteins, metabolites, and clinical factors. This approach manages challenges like high dimensionality, small sample sizes, and data heterogeneity. By applying different feature extraction and fusion strategies, notably late fusion models, the effectiveness of integrating diverse data types was demonstrated. Late fusion models consistently outperformed single-modality approaches in TCGA lung, breast, and pan-cancer datasets, offering higher accuracy and robustness. This research highlights the potential of comprehensive multimodal data integration in precision oncology to improve survival predictions for cancer patients. The study provides a reusable pipeline for the research community, suggesting future work on larger cohorts.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: All authors are or were employees of AstraZeneca at the time this work was performed and may have stock ownership, options, or interests in the company.
Figures


References
-
- Baltrusaitis, T., Ahuja, C. & Morency, L. P. Multimodal machine learning: a survey and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell.41, 423–443 (2019). - PubMed
-
- Huang, Y. et al. In Adv Neural Inf Process Syst. 34 (eds. M. Ranzato et al.) 1–13 (Neural Information Processing Systems Foundation, 2021).
LinkOut - more resources
Full Text Sources
Miscellaneous

