Near-complete Middle Eastern genomes refine autozygosity and enhance disease-causing and population-specific variant discovery
- PMID: 40325133
- PMCID: PMC12081309
- DOI: 10.1038/s41588-025-02173-7
Near-complete Middle Eastern genomes refine autozygosity and enhance disease-causing and population-specific variant discovery
Abstract
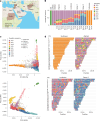
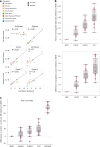
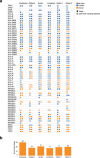
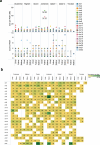
Advances in long-read sequencing have enabled routine complete assembly of human genomes, but much remains to be done to represent broader populations and show impact on disease-gene discovery. Here, we report highly accurate, near-complete and phased genomes from six Middle Eastern (ME) family trios (n = 18) with neurodevelopmental conditions, representing ancestries from Sudan, Jordan, Syria, Qatar and Afghanistan. These genomes revealed 42.2 Mb of new sequence (13.8% impacting known genes), 75 new HLA/KIR alleles and strong signals of inbreeding, with ROH covering up to one-third of chromosomes 6 and 12 in one individual. Using assembly-based variant calling, we identified 23 de novo and recessive variants as strong candidates for causing previously unresolved symptoms in the probands. The ME genomes revealed unique variation relative to existing references, showing enhanced mappability and variant calling. These results underscore the value of de novo assembly for disease variant discovery and the need for sampled ME-specific references to better characterize population-relevant variation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: E.E.E. is a scientific advisory board member of Variant Bio. The other authors declare no competing interests.
Figures






References
-
- Seo, J. S. et al. De novo assembly and phasing of a Korean human genome. Nature538, 243–247 (2016). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

