In Vitro Expansion and Transduction of Primary NK Cells Using Feeder Cells Expressing Costimulatory Molecules and IL-21
- PMID: 40325497
- PMCID: PMC12210053
- DOI: 10.1111/cas.70090
In Vitro Expansion and Transduction of Primary NK Cells Using Feeder Cells Expressing Costimulatory Molecules and IL-21
Abstract
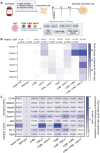
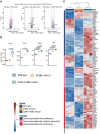
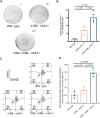
Natural Killer (NK) cells are an important population of the immune system, and NK cell-based therapy has shown great potential in the treatment of cancers. However, to apply NK cells clinically, producing a large number of cells with high cytotoxicity remains a challenge. Current strategies focus on employing different irradiated feeder cells to stimulate NK expansion, maturation, and cytotoxicity. While co-stimulatory signals play critical roles in promoting NK cell proliferation and activating their functions, the exploitation of these signals for expanding NK cells has not been fully explored. To identify the optimal engineered feeder cells for expanding umbilical cord blood-derived NK cells, we generated different feeder cells expressing the co-stimulatory molecules CD80, 4-1BBL, or membrane-bound IL-21 (mbIL21). We then evaluated the transduction efficacy of a chimeric antigen receptor (CAR) construct into expanded NK cells using various lentiviral vectors. Our results showed that CD80, in combination with 4-1BBL and mbIL21, induced the highest expansion of NK cells from cord blood. The expanded NK cells displayed higher cytotoxicity toward target cells compared to T cells following CAR transduction using BaEV lentivirus.
Keywords: BaEV lentivirus; CAR‐NK; CD80; CD80‐41BBL‐mbIL21 K562 cells; umbilical cord blood‐derived NK cells.
© 2025 The Author(s). Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
Hoai‐Nghia Nguyen and Le Son Tran are affiliated with Gene Solutions JSC. Other authors declare no conflicts of interest.
Figures


References
-
- Wolf N. K., Kissiov D. U., and Raulet D. H., “Roles of Natural Killer Cells in Immunity to Cancer, and Applications to Immunotherapy,” Nature Reviews. Immunology 23, no. 2 (2023): 90–105. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

