Transition Temperature-Guided Design of Lipid Nanoparticles for Effective mRNA Delivery
- PMID: 40325908
- PMCID: PMC12086761
- DOI: 10.1021/acsami.5c06464
Transition Temperature-Guided Design of Lipid Nanoparticles for Effective mRNA Delivery
Abstract
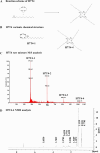
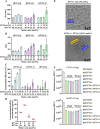
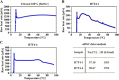
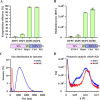
Lipid nanoparticles (LNPs) are promising mRNA delivery vehicles due to their biocompatibility and tunable characteristics. While current rational design approaches focus on ionizable lipids' pKa and zeta potential to optimize mRNA encapsulation and endosomal escape, the selection of helper lipids remains largely empirical. We propose that the lipid transition temperature (Tm), marking the shift from the gel to the liquid crystalline phase, can guide rational helper lipid selection. Through screening 54 ionizable lipids, we identified H7T4, which displayed favorable physicochemical properties when combined with its tail variants but exhibited poor transfection efficiency. Using nano differential scanning calorimetry (nDSC) and biological small-angle X-ray scattering (BioSAXS), we found that lowering the system's Tm by combining H7T4 (high transition temperature) with a low-transition-temperature helper lipid such as 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) significantly enhanced mRNA cellular uptake both in vitro and in vivo. These findings establish Tm as a crucial parameter for a rational LNP design.
Keywords: SAXS; ionizable lipid; lipid nanoparticle; mRNA; nDSC.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Sarnelli S.; Cardamone M.; Reverchon E.; Baldino L. Lipid-Based Nanoparticles for Nucleic Acids Delivery. Phys. Sci. Rev. 2025, 10, 317–338. 10.1515/psr-2025-0001. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

