Lipid Scrambling Pathways in the Sec61 Translocon Complex
- PMID: 40325981
- PMCID: PMC12082634
- DOI: 10.1021/jacs.4c11142
Lipid Scrambling Pathways in the Sec61 Translocon Complex
Abstract
Cellular homeostasis depends on the rapid, ATP-independent translocation of newly synthesized lipids across the endoplasmic reticulum (ER) membrane. Lipid translocation is facilitated by membrane proteins known as scramblases, a few of which have recently been identified in the ER. Our previous structure of the translocon-associated protein (TRAP) bound to the Sec61 translocation channel revealed local membrane thinning, suggesting that the Sec61/TRAP complex might be involved in lipid scrambling. Using complementary fluorescence spectroscopy assays, we detected nonselective scrambling by reconstituted translocon complexes. This activity was unaffected by Sec61 inhibitors that block its lateral gate, suggesting a second lipid scrambling pathway within the complex. Molecular dynamics simulations indicate that the trimeric TRAP subunit forms this alternative route, facilitating lipid translocation via a "credit card" mechanism, using a crevice lined with polar residues to shield lipid head groups from the hydrophobic membrane interior. Kinetic and thermodynamic analyses confirmed that local membrane thinning enhances scrambling efficiency and that both Sec61 and TRAP scramble phosphatidylcholine faster than phosphatidylethanolamine and phosphatidylserine, reflecting the intrinsic lipid flip-flop tendencies of these lipid species. As the Sec61 scrambling site lies in the lateral gate region, it is likely inaccessible during protein translocation, in line with our experiments on Sec61-inhibited samples. Hence, our findings suggest that the metazoan-specific trimeric TRAP bundle is a viable candidate for lipid scrambling activity that is insensitive to the functional state of the translocon.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

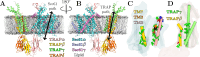

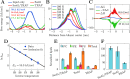
 , from which the activation energy EA is obtained. The error bars show relative
error, yet they are not visible for most of the data points. E) Lipid
headgroup selectivity of the scrambling activity of Sec61, TRAP, and
Sec61/TRAP complexes. Bars show the mean and standard error for the
number of total scrambled lipids extracted from the five 20 μs-long
replica simulations of the multicomponent membrane (Set 2 in Table 1). The membranes contain
equal amounts of PC, PE, and PS. F) Scrambling of POPC by Sec61, TRAP,
and Sec61/TRAP complexes in the single-component membrane (Set 1 in Table 1). Bars show the mean
and standard error for the number of total scrambled lipids extracted
from the five 20 μs-long replica simulations.
, from which the activation energy EA is obtained. The error bars show relative
error, yet they are not visible for most of the data points. E) Lipid
headgroup selectivity of the scrambling activity of Sec61, TRAP, and
Sec61/TRAP complexes. Bars show the mean and standard error for the
number of total scrambled lipids extracted from the five 20 μs-long
replica simulations of the multicomponent membrane (Set 2 in Table 1). The membranes contain
equal amounts of PC, PE, and PS. F) Scrambling of POPC by Sec61, TRAP,
and Sec61/TRAP complexes in the single-component membrane (Set 1 in Table 1). Bars show the mean
and standard error for the number of total scrambled lipids extracted
from the five 20 μs-long replica simulations.References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

