Tumor suppressors in Sox2-mediated lung cancers promote distinct cell-intrinsic and immunologic remodeling
- PMID: 40327401
- PMCID: PMC12220946
- DOI: 10.1172/jci.insight.171364
Tumor suppressors in Sox2-mediated lung cancers promote distinct cell-intrinsic and immunologic remodeling
Abstract
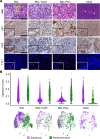
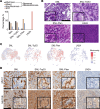
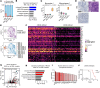
Non-small cell lung cancer (NSCLC) largely consists of lung squamous carcinoma (LUSC) and lung adenocarcinoma (LUAD). Alterations in the tumor protein p53 (TP53) and phosphatase and tensin homolog (PTEN) tumor suppressors are common in both subtypes, but their relationship with SOX2 is poorly understood. We deleted Trp53 or Pten in a C57BL/6 Sox2hi Nkx2-1-/- Lkb1-/- (SNL) genetic background and generated a highly metastatic LUSC cell line (LN2A; derived from a Sox2hi mouse model, followed by Trp53, Pten, and cyclin dependent kinase inhibitor 2A [Cdkn2a] deletion). Histologic and single-cell RNA-Seq analyses corroborated that SNL mice developed mixed tumors with both LUAD and LUSC histopathology while SNL-Trp53 and SNL-Pten mice developed LUAD and LN2A tumors that retained LUSC morphology. Compared with SNL mice, additional loss of Trp53 or Pten resulted in significantly reduced survival, increased tumor burden, and altered tumor mucin composition. We identified a subcluster of CD38+ tumor-associated inflammatory monocytes in the LN2A model that was significantly enriched for activation of the classical and alternative complement pathways. Complement factor B (CFB) is associated with poor survival in patients with LUSC, and we observed the LN2A model had significantly improved survival on a Cfb-/- background. Our findings demonstrate a cooperative role of Trp53 and Pten tumor suppressors in Sox2-mediated NSCLC tumor progression, mucin production, and remodeling of the immune tumor microenvironment.
Keywords: Genetics; Innate immunity; Mouse models; Oncology.
Figures


References
-
- American Cancer Society. Global Cancer Facts & Figures 5th Edition. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-... Updated 2024. Accessed May 23, 2025.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

