The Integrated Stress Response Pathway Coordinates Translational Control of Multiple Immune Checkpoints in Lung Cancer
- PMID: 40327600
- PMCID: PMC12260515
- DOI: 10.1158/0008-5472.CAN-24-3844
The Integrated Stress Response Pathway Coordinates Translational Control of Multiple Immune Checkpoints in Lung Cancer
Abstract
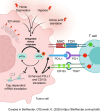
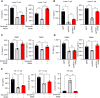
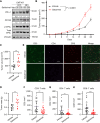
The integrated stress response (ISR) is an adaptive pathway hijacked by cancer cells to survive cellular stresses in the tumor microenvironment. ISR activation potently induces PD-L1, leading to suppression of antitumor immunity. In this study, we sought to uncover additional immune checkpoint proteins regulated by the ISR to elucidate mechanisms of tumor immune escape. The ISR coordinately induced cluster of differentiation 155 (CD155) and PD-L1, enhancing translation of both immune checkpoint proteins through bypass of inhibitory upstream open reading frames in their 5' untranslated regions. Analysis of primary human lung tumors identified a significant correlation between expression of PD-L1 and CD155. ISR activation accelerated tumorigenesis and inhibited T-cell function, which could be overcome by combining PD-1 and TIGIT blockade with the ISR inhibitor ISRIB. This study uncovers a mechanism by which two immune checkpoint proteins are coordinately regulated and suggests a therapeutic strategy for patients with lung cancer.
Significance: The integrated stress response represents a targetable axis to improve the efficacy of immunotherapy in lung cancer by inhibiting the coordinated translational regulation of the PD-L1/PD-1 and CD155/TIGIT immune checkpoint pathways.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
S. Thomas-Jardin reports grants from the NIH/NCI, Cancer Prevention Research Institute of Texas (CPRIT), NIH–University of Texas SPORE in Lung Cancer, NIH–UTSW Whole Brain imaging Core, NIH/NCI during the conduct of the study. A. Arce reports grants from the NCI, CPRIT, Welch Foundation, V Foundation, and Department of Defense during the conduct of the study. C. Lewis reports other support from Guardant Health, Nurix Therapeutics, Caleo Biotechnologies, and Iovance Biotherapeutics outside the submitted work. B.M. Evers reports grants from the NCI, CPRIT, Welch Foundation, V Foundation, and Department of Defense during the conduct of the study. I.I. Wistuba reports grants and personal fees from Amgen, Johnson & Johnson, Genentech/Roche, Bristol Myers Squibb, Bayer, AstraZeneca, Pfizer, Merck, and Novartis, personal fees from Flame, Guardant Health, Sanofi, Regeneron, Daiichi Sankyo, Jansen, G1 Therapeutics, Merus, AbbVie, Catalyst Therapeutics, Boehringer Ingelheim, and Oncocyte, and grants from Medimmune, Karus, 4D, Iovance Biotherapeutics, Adaptimmune, EMD Serono, Adaptive, Takeda, Akoya, Dava Oncology, Physician Education Resource, Suzhou Liangyihui Network Technology, and Platform Health outside the submitted work. J.D. Minna reports grants from the NCI and personal fees from the NIH and UTSW Medical Center during the conduct of the study. K.A. O’Donnell reports grants from the NCI, CPRIT, Welch Foundation, V Foundation, Department of Defense, and NCI during the conduct of the study and serves on the Scientific Advisory Board of the Lung Cancer Research Foundation. No disclosures were reported by the other authors.
Figures





Update of
-
Coordinated translational control of multiple immune checkpoints by the integrated stress response pathway in lung cancer.bioRxiv [Preprint]. 2024 Oct 28:2024.10.23.619897. doi: 10.1101/2024.10.23.619897. bioRxiv. 2024. Update in: Cancer Res. 2025 Jul 15;85(14):2574-2590. doi: 10.1158/0008-5472.CAN-24-3844. PMID: 39554171 Free PMC article. Updated. Preprint.
References
-
- Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 2002;8:793–800. - PubMed
-
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006;439:682–7. - PubMed
MeSH terms
Substances
Grants and funding
- I-1881/Welch Foundation (The Welch Foundation)
- P50CA70907/National Cancer Institute (NCI)
- R01 CA207763/CA/NCI NIH HHS/United States
- R01CA207763/National Cancer Institute (NCI)
- P30 CA142543/CA/NCI NIH HHS/United States
- LC190249/U.S. Department of Defense (DOD)
- RP200327/Cancer Prevention and Research Institute of Texas (CPRIT)
- R01CA273585/National Cancer Institute (NCI)
- P30CA142543/National Cancer Institute (NCI)
- R01 CA273585/CA/NCI NIH HHS/United States
- F32CA274982/National Cancer Institute (NCI)
- RP250391/Cancer Prevention and Research Institute of Texas (CPRIT)
- S10 OD032267/OD/NIH HHS/United States
- T2021-011/V Foundation for Cancer Research (VFCR)
- IHP24002/Melanoma Research Foundation (MRF)
- P50 CA070907/CA/NCI NIH HHS/United States
- F32 CA274982/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

