Custom-made medium approach for effective enrichment and isolation of chemolithotrophic iron-oxidizing bacteria
- PMID: 40328454
- PMCID: PMC12089753
- DOI: 10.1093/femsec/fiaf051
Custom-made medium approach for effective enrichment and isolation of chemolithotrophic iron-oxidizing bacteria
Abstract
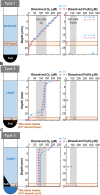
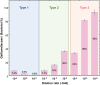
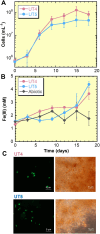
Chemolithotrophic neutrophilic iron (Fe)-oxidizing bacteria, which mainly belong to the family Gallionellaceae, universally prevail in terrestrial environments changing Fe cycling. However, they are typically recognized as difficult-to-culture microbes. Despite efforts, there are few Fe(II)-oxidizing lithotroph isolates; hence, their physiological and ecological knowledge remains limited. This limitation is largely owing to difficulties in their cultivation, and we hypothesize that the difficulty exists because substrate and mineral concentrations in the cultivation medium are not tuned to a specific environmental condition under which these organisms live. To address this hypothesis, this study proposes a novel custom-made medium approach for chemolithotrophic Fe(II)-oxidizing bacteria; a method that manipulates medium components through diligent analysis of field environment. A new custom-made medium simulating energy substrates and nutrients under the field condition was prepared by modifying both chemical composition and physical setup in the glass-tube medium. In particular, the modification of the physical setup in the tube had a significant effect on adjusting dissolved Fe(II) and O2 concentrations to the field environment. Using the medium, Gallionellaceae members were successfully enriched and a new Gallionellaceae species was isolated from a natural hot spring site. Compared with conventional medium, the custom-made medium has significantly higher ability in enriching Gallionellaceae members.
Keywords: Gallionellaceae; custom-made medium; enrichment; iron-oxidizing bacteria; isolation.
© The Author(s) 2025. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
All authors declared no conflicts of interest.
Figures




References
-
- Appelo CAJ, Postma D. Geochemistry, Groundwater and Pollution(2nd ed.). London: CRC Press, 2005.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

