Unlocking Lactonase Enzymes as Biocatalysts for the Deracemisation of Chiral γ-Thiolactones
- PMID: 40328665
- PMCID: PMC12258673
- DOI: 10.1002/anie.202505032
Unlocking Lactonase Enzymes as Biocatalysts for the Deracemisation of Chiral γ-Thiolactones
Abstract
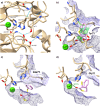
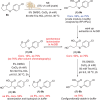
Lactonases, a class of metalloenzymes that exhibit catalytic promiscuity, have been extensively studied from a biological perspective, yet their application as biocatalysts remains underexplored. In this study, we disclose the biocatalytic activity of lactonase enzymes in the hydrolysis and deracemisation of chiral C3-substituted-γ-thiolactones and the asymmetric synthesis of γ-thio-α-substituted-carboxylic acids. The thiolactonase activity of lactonases from different protein superfamilies was investigated. The biocatalyst GcL, from the metallo-β-lactamase-like lactonase family, catalysed the enzymatic kinetic resolution (EKR) of homocysteine (Hcy) thiolactones with excellent enantioselectivity (E-value up to 136), yielding enantioenriched Hcy thiolactones and γ-thio-α-amino-carboxylic acids with high ees. Additionally, the biocatalyst N9 Y71G, a rationally engineered variant of the reconstructed ancestral paraoxonase enzyme N9, catalysed the dynamic kinetic resolution (DKR) of C3-thio-γ-thiolactones, yielding γ-thio-α-thio-carboxylic acids in enantioselective manner with high ees (up to >99%) and yields (up to >99%). Insights on the mechanism and the stereoselectivity of the lactonase biocatalysts were gained through computational and site-directed mutagenesis studies.
Keywords: Biocatalysis; Kinetic resolution; Lactonase; Sulphur compounds; Thiolactones.
© 2025 The Author(s). Angewandte Chemie International Edition published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

